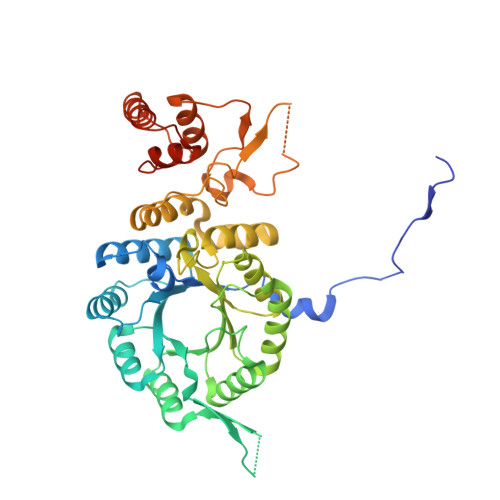
Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis.
Bulfer, S.L., Scott, E.M., Couture, J.F., Pillus, L., Trievel, R.C.(2009) J Biol Chem 284: 35769-35780
- PubMed: 19776021
- DOI: https://doi.org/10.1074/jbc.M109.046821
- Primary Citation of Related Structures:
3IVS, 3IVT, 3IVU - PubMed Abstract:
Homocitrate synthase (HCS) catalyzes the first and committed step in lysine biosynthesis in many fungi and certain Archaea and is a potential target for antifungal drugs. Here we report the crystal structure of the HCS apoenzyme from Schizosaccharomyces pombe and two distinct structures of the enzyme in complex with the substrate 2-oxoglutarate (2-OG). The structures reveal that HCS forms an intertwined homodimer stabilized by domain-swapping between the N- and C-terminal domains of each monomer. The N-terminal catalytic domain is composed of a TIM barrel fold in which 2-OG binds via hydrogen bonds and coordination to the active site divalent metal ion, whereas the C-terminal domain is composed of mixed alpha/beta topology. In the structures of the HCS apoenzyme and one of the 2-OG binary complexes, a lid motif from the C-terminal domain occludes the entrance to the active site of the neighboring monomer, whereas in the second 2-OG complex the lid is disordered, suggesting that it regulates substrate access to the active site through its apparent flexibility. Mutations of the active site residues involved in 2-OG binding or implicated in acid-base catalysis impair or abolish activity in vitro and in vivo. Together, these results yield new insights into the structure and catalytic mechanism of HCSs and furnish a platform for developing HCS-selective inhibitors.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, Ann Arbor, Michigan 48109, USA.