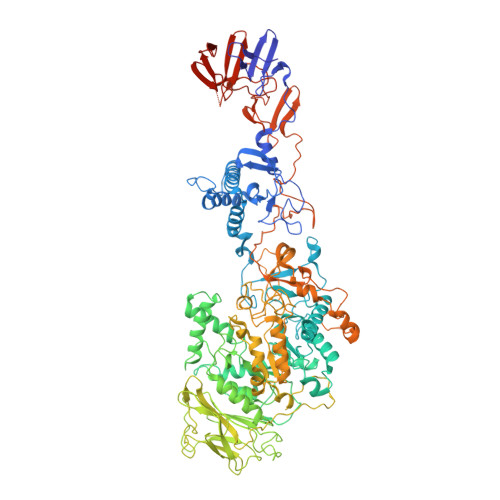
Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Vujicic-Zagar, A., Pijning, T., Kralj, S., Lopez, C.A., Eeuwema, W., Dijkhuizen, L., Dijkstra, B.W.(2010) Proc Natl Acad Sci U S A 107: 21406-21411
- PubMed: 21118988
- DOI: https://doi.org/10.1073/pnas.1007531107
- Primary Citation of Related Structures:
3HZ3, 3KLK, 3KLL - PubMed Abstract:
Glucansucrases are large enzymes belonging to glycoside hydrolase family 70, which catalyze the cleavage of sucrose into fructose and glucose, with the concomitant transfer of the glucose residue to a growing α-glucan polymer. Among others, plaque-forming oral bacteria secrete these enzymes to produce α-glucans, which facilitate the adhesion of the bacteria to the tooth enamel. We determined the crystal structure of a fully active, 1,031-residue fragment encompassing the catalytic and C-terminal domains of GTF180 from Lactobacillus reuteri 180, both in the native state, and in complexes with sucrose and maltose. These structures show that the enzyme has an α-amylase-like (β/α)(8)-barrel catalytic domain that is circularly permuted compared to the catalytic domains of members of glycoside hydrolase families 13 and 77, which belong to the same GH-H superfamily. In contrast to previous suggestions, the enzyme has only one active site and one nucleophilic residue. Surprisingly, in GTF180 the peptide chain follows a "U"-path, such that four of the five domains are made up from discontiguous N- and C-terminal stretches of the peptide chain. Finally, the structures give insight into the factors that determine the different linkage types in the polymeric product.
Organizational Affiliation:
Laboratory of Biophysical Chemistry, University of Groningen, Nijenborgh 4, 9747 AG Groningen, The Netherlands.