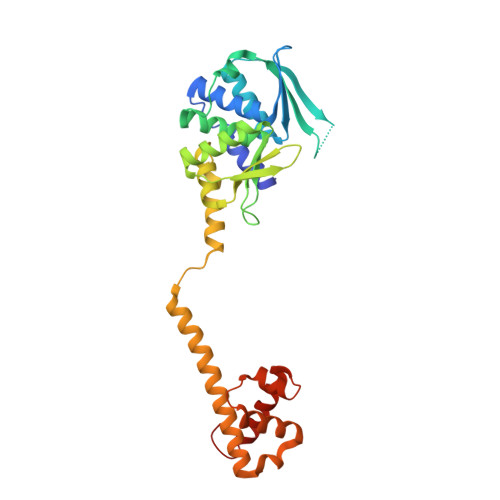
The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease
Kaiser, B.K., Clifton, M.C., Shen, B.W., Stoddard, B.L.(2009) Structure 17: 1368-1376
- PubMed: 19836336
- DOI: https://doi.org/10.1016/j.str.2009.08.008
- Primary Citation of Related Structures:
3HYI, 3HYJ - PubMed Abstract:
Proteins of the DUF199 family, present in all Gram-positive bacteria and best characterized by the WhiA sporulation control factor in Streptomyces coelicolor, are thought to act as genetic regulators. The crystal structure of the DUF199/WhiA protein from Thermatoga maritima demonstrates that these proteins possess a bipartite structure, in which a degenerate N-terminal LAGLIDADG homing endonuclease (LHE) scaffold is tethered to a C-terminal helix-turn-helix (HTH) domain. The LHE domain has lost those residues critical for metal binding and catalysis, and also displays an extensively altered DNA-binding surface as compared with homing endonucleases. The HTH domain most closely resembles related regions of several bacterial sigma70 factors that bind the -35 regions of bacterial promoters. The structure illustrates how an invasive element might be transformed during evolution into a larger assemblage of protein folds that can participate in the regulation of a complex biological pathway.
Organizational Affiliation:
Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue N., Seattle, WA 98006, USA.