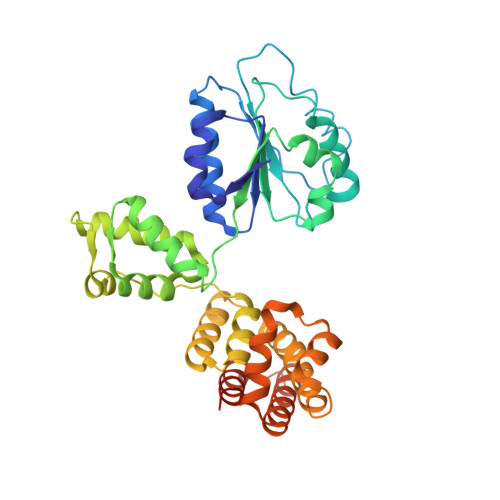
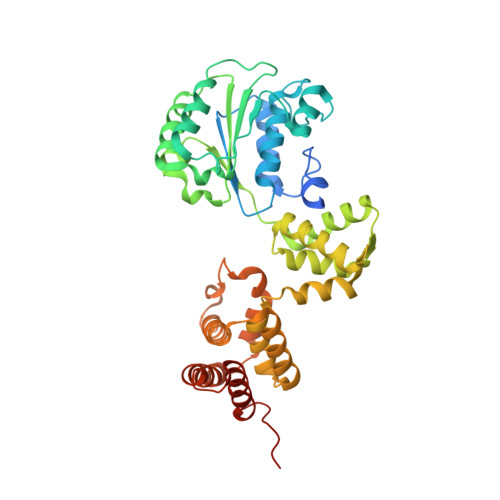
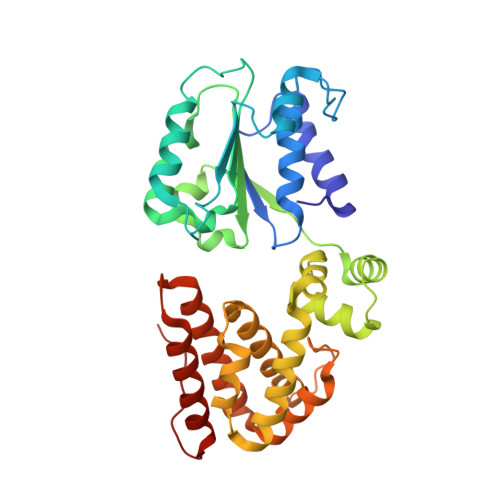
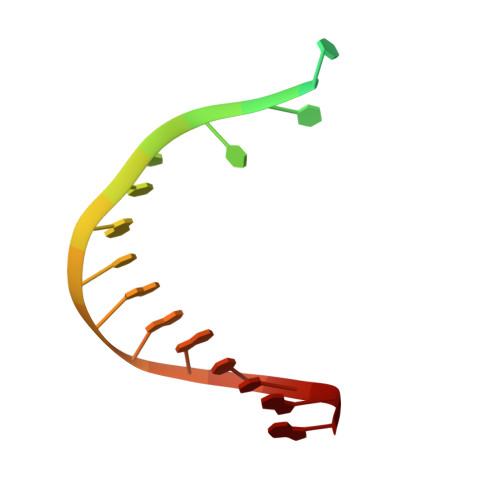
The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Simonetta, K.R., Kazmirski, S.L., Goedken, E.R., Cantor, A.J., Kelch, B.A., McNally, R., Seyedin, S.N., Makino, D.L., O'Donnell, M., Kuriyan, J.(2009) Cell 137: 659-671
- PubMed: 19450514
- DOI: https://doi.org/10.1016/j.cell.2009.03.044
- Primary Citation of Related Structures:
3GLF, 3GLG, 3GLH, 3GLI - PubMed Abstract:
Clamp loaders load sliding clamps onto primer-template DNA. The structure of the E. coli clamp loader bound to DNA reveals the formation of an ATP-dependent spiral of ATPase domains that tracks only the template strand, allowing recognition of both RNA and DNA primers. Unlike hexameric helicases, in which DNA translocation requires distinct conformations of the ATPase domains, the clamp loader spiral is symmetric and is set up to trigger release upon DNA recognition. Specificity for primed DNA arises from blockage of the end of the primer and accommodation of the emerging template along a surface groove. A related structure reveals how the psi protein, essential for coupling the clamp loader to single-stranded DNA-binding protein (SSB), binds to the clamp loader. By stabilizing a conformation of the clamp loader that is consistent with the ATPase spiral observed upon DNA binding, psi binding promotes the clamp-loading activity of the complex.
Organizational Affiliation:
Department of Molecular and Cell Biology, Howard Hughes Medical Institute, University of California, Berkeley, Berkeley, CA 94720, USA.