Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA.
Agarwal, S., Hong, D., Desai, N.K., Sazinsky, M.H., Arguello, J.M., Rosenzweig, A.C.(2010) Proteins 78: 2450-2458
- PubMed: 20602459
- DOI: https://doi.org/10.1002/prot.22753
- Primary Citation of Related Structures:
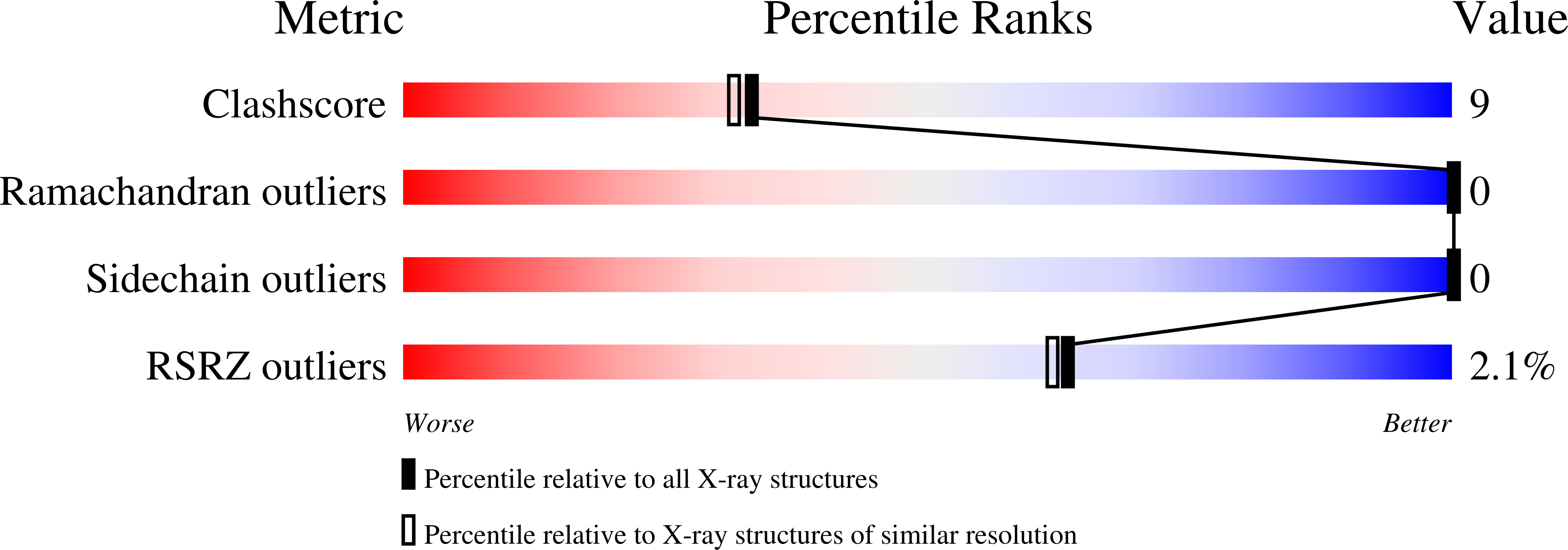
3FRY - PubMed Abstract:
The Cu(+)-ATPase CopA from Archaeoglobus fulgidus belongs to the P(1B) family of the P-type ATPases. These integral membrane proteins couple the energy of ATP hydrolysis to heavy metal ion translocation across membranes. A defining feature of P(1B-1)-type ATPases is the presence of soluble metal binding domains at the N-terminus (N-MBDs). The N-MBDs exhibit a conserved ferredoxin-like fold, similar to that of soluble copper chaperones, and bind metal ions via a conserved CXXC motif. The N-MBDs enable Cu(+) regulation of turnover rates apparently through Cu-sensitive interactions with catalytic domains. A. fulgidus CopA is unusual in that it contains both an N-terminal MBD and a C-terminal MBD (C-MBD). The functional role of the unique C-MBD has not been established. Here, we report the crystal structure of the apo, oxidized C-MBD to 2.0 A resolution. In the structure, two C-MBD monomers form a domain-swapped dimer, which has not been observed previously for similar domains. In addition, the interaction of the C-MBD with the other cytoplasmic domains of CopA, the ATP binding domain (ATPBD) and actuator domain (A-domain), has been investigated. Interestingly, the C-MBD interacts specifically with both of these domains, independent of the presence of Cu(+) or nucleotides. These data reinforce the uniqueness of the C-MBD and suggest a distinct structural role for the C-MBD in CopA transport.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, Evanston, Illinois 60208, USA.