Functional annotation and three-dimensional structure of Dr0930 from Deinococcus radiodurans, a close relative of phosphotriesterase in the amidohydrolase superfamily.
Xiang, D.F., Kolb, P., Fedorov, A.A., Meier, M.M., Fedorov, L.V., Nguyen, T.T., Sterner, R., Almo, S.C., Shoichet, B.K., Raushel, F.M.(2009) Biochemistry 48: 2237-2247
- PubMed: 19159332
- DOI: https://doi.org/10.1021/bi802274f
- Primary Citation of Related Structures:
3FDK - PubMed Abstract:
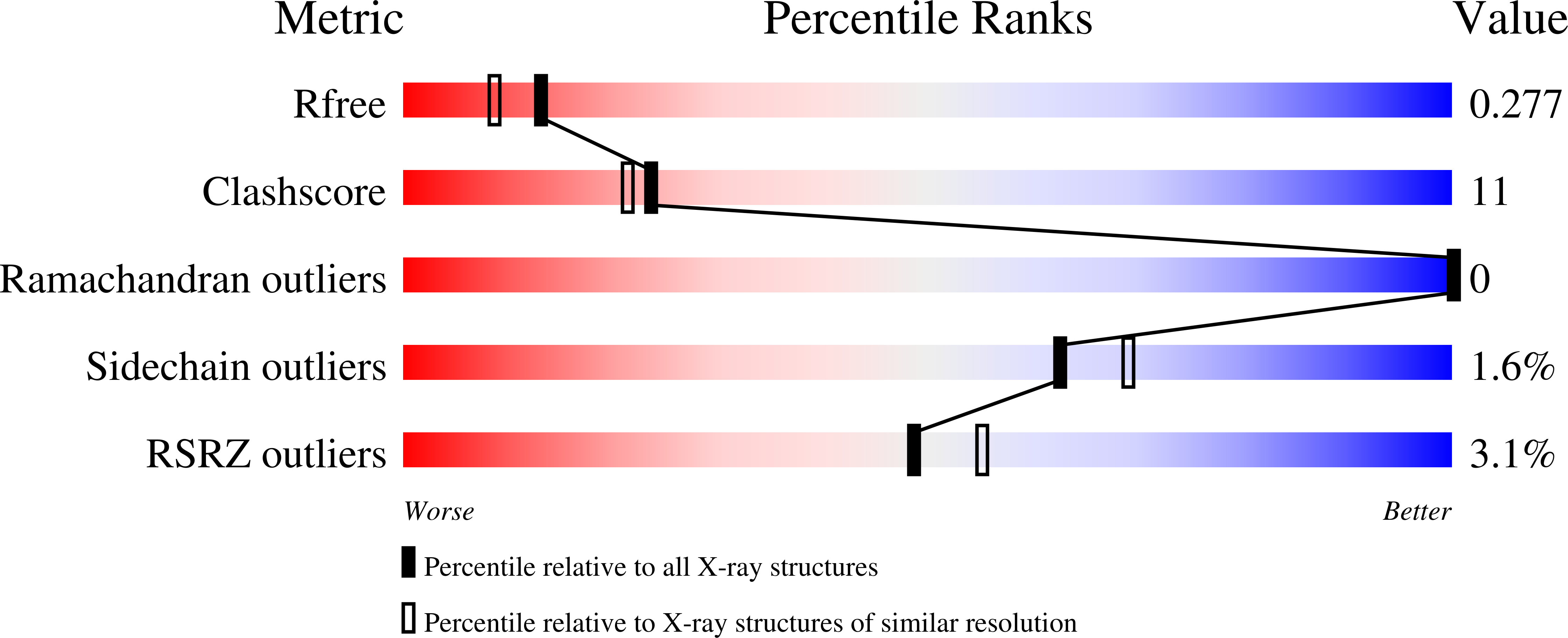
Dr0930, a member of the amidohydrolase superfamily in Deinococcus radiodurans, was cloned, expressed, and purified to homogeneity. The enzyme crystallized in the space group P3121, and the structure was determined to a resolution of 2.1 A. The protein folds as a (beta/alpha)7beta-barrel, and a binuclear metal center is found at the C-terminal end of the beta-barrel. The purified protein contains a mixture of zinc and iron and is intensely purple at high concentrations. The purple color was determined to be due to a charge transfer complex between iron in the beta-metal position and Tyr-97. Mutation of Tyr-97 to phenylalanine or complexation of the metal center with manganese abolished the absorbance in the visible region of the spectrum. Computational docking was used to predict potential substrates for this previously unannotated protein. The enzyme was found to catalyze the hydrolysis of delta- and gamma-lactones with an alkyl substitution at the carbon adjacent to the ring oxygen. The best substrate was delta-nonanoic lactone with a kcat/Km of 1.6 x 10(6) M-1 s-1. Dr0930 was also found to catalyze the very slow hydrolysis of paraoxon with values of kcat and kcat/Km of 0.07 min-1 and 0.8 M-1 s-1, respectively. The amino acid sequence identity to the phosphotriesterase (PTE) from Pseudomonas diminuta is 30%. The eight substrate specificity loops were transplanted from PTE to Dr0930, but no phosphotriesterase activity could be detected in the chimeric PTE-Dr0930 hybrid. Mutation of Phe-26 and Cys-72 in Dr0930 to residues found in the active site of PTE enhanced the kinetic constants for the hydrolysis of paraoxon. The F26G/C72I mutant catalyzed the hydrolysis of paraoxon with a kcat of 1.14 min-1, an increase of 16-fold over the wild-type enzyme. These results support previous proposals that phosphotriesterase activity evolved from an ancestral parent enzyme possessing lactonase activity.
Organizational Affiliation:
Department of Chemistry, Texas A&M University, P.O. Box 30012, College Station, Texas 77842-3012, USA.