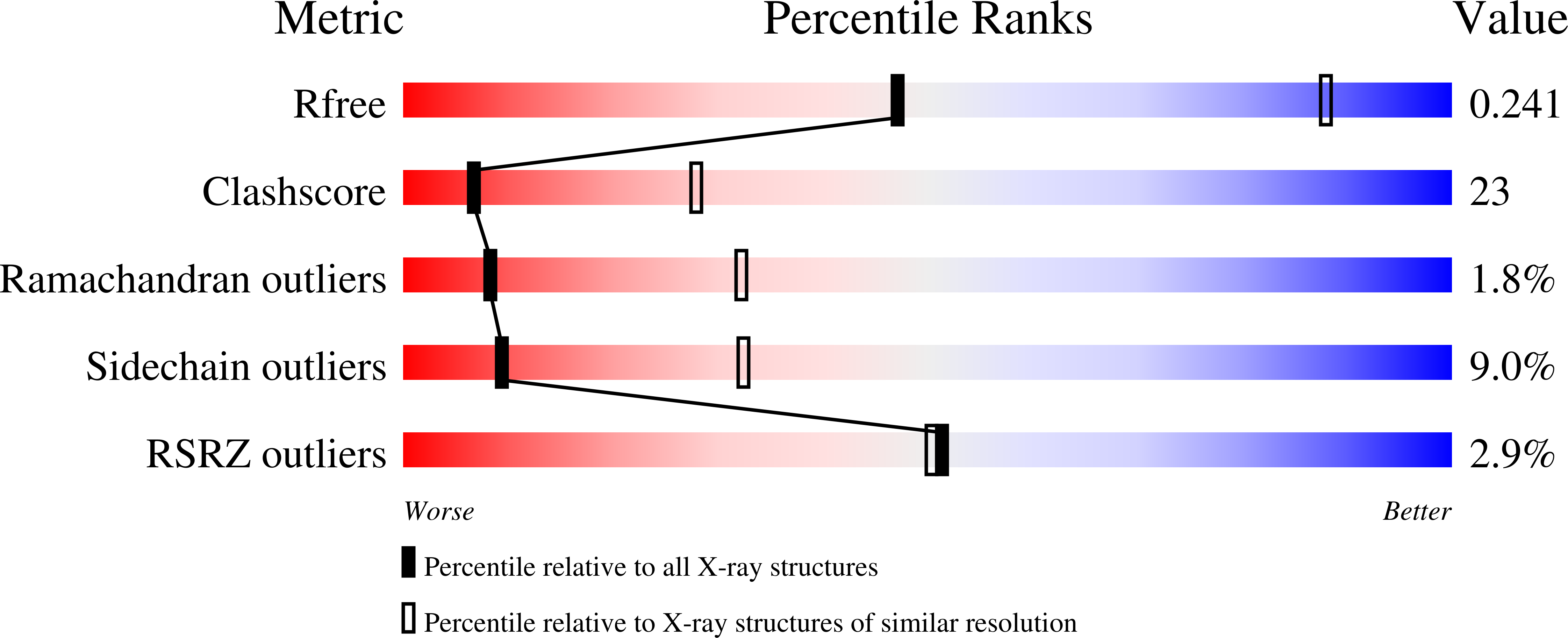
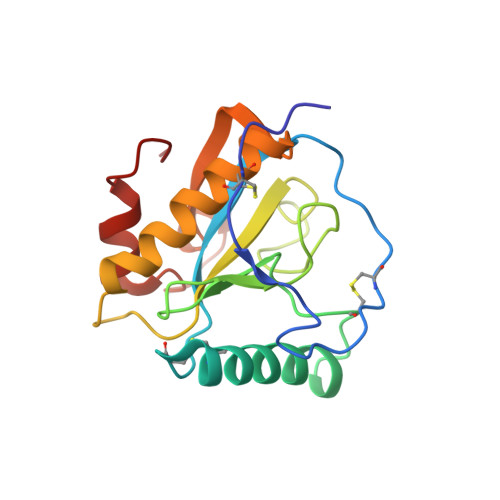
Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution
Balaji, K., Sharma, P., Singh, N., Sinha, M., Bhushan, A., Kaur, P., Sharma, S., Singh, T.P.To be published.