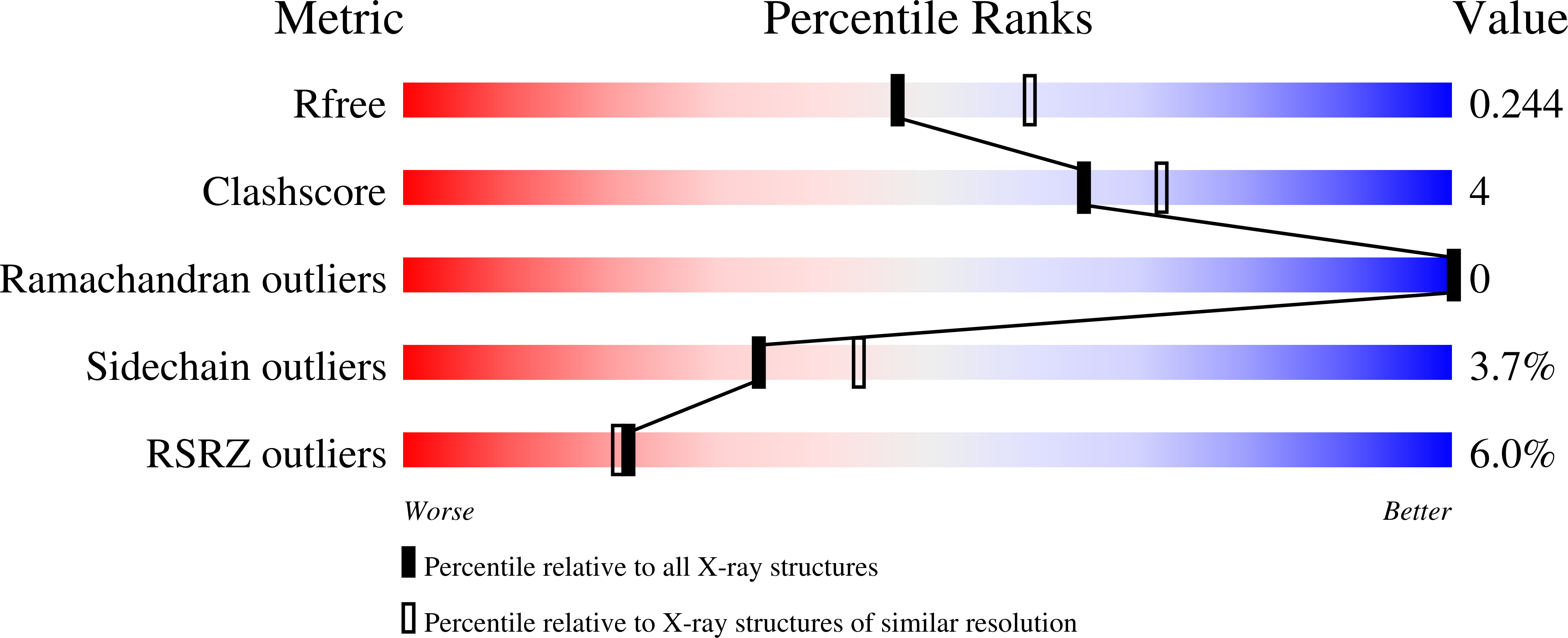
Liprin-mediated large signaling complex organization revealed by the liprin-alpha/CASK and liprin-alpha/liprin-beta complex structures
Wei, Z., Zheng, S., Spangler, S.A., Yu, C., Hoogenraad, C.C., Zhang, M.(2011) Mol Cell 43: 586-598
- PubMed: 21855798
- DOI: https://doi.org/10.1016/j.molcel.2011.07.021
- Primary Citation of Related Structures:
3TAC, 3TAD - PubMed Abstract:
Liprins are highly conserved scaffold proteins that regulate cell adhesion, cell migration, and synapse development by binding to diverse target proteins. The molecular basis governing liprin/target interactions is poorly understood. The liprin-α2/CASK complex structure solved here reveals that the three SAM domains of liprin-α form an integrated supramodule that binds to the CASK kinase-like domain. As supported by biochemical and cellular studies, the interaction between liprin-α and CASK is unique to vertebrates, implying that the liprin-α/CASK interaction is likely to regulate higher-order brain functions in mammals. Consistently, we demonstrate that three recently identified X-linked mental retardation mutants of CASK are defective in binding to liprin-α. We also solved the liprin-α/liprin-β SAM domain complex structure, which uncovers the mechanism underlying liprin heterodimerizaion. Finally, formation of the CASK/liprin-α/liprin-β ternary complex suggests that liprins can mediate assembly of target proteins into large protein complexes capable of regulating numerous cellular activities.
Organizational Affiliation:
Division of Life Science, State Key Laboratory of Molecular Neuroscience, Molecular Neuroscience Center, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China.