A 1.5 A resolution X-ray structure of the catalytic module of Caldicellulosiruptor bescii family 3 pectate lyase.
Alahuhta, M., Chandrayan, P., Kataeva, I., Adams, M.W., Himmel, M.E., Lunin, V.V.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1498-1500
- PubMed: 22139151
- DOI: https://doi.org/10.1107/S1744309111038449
- Primary Citation of Related Structures:
3T9G - PubMed Abstract:
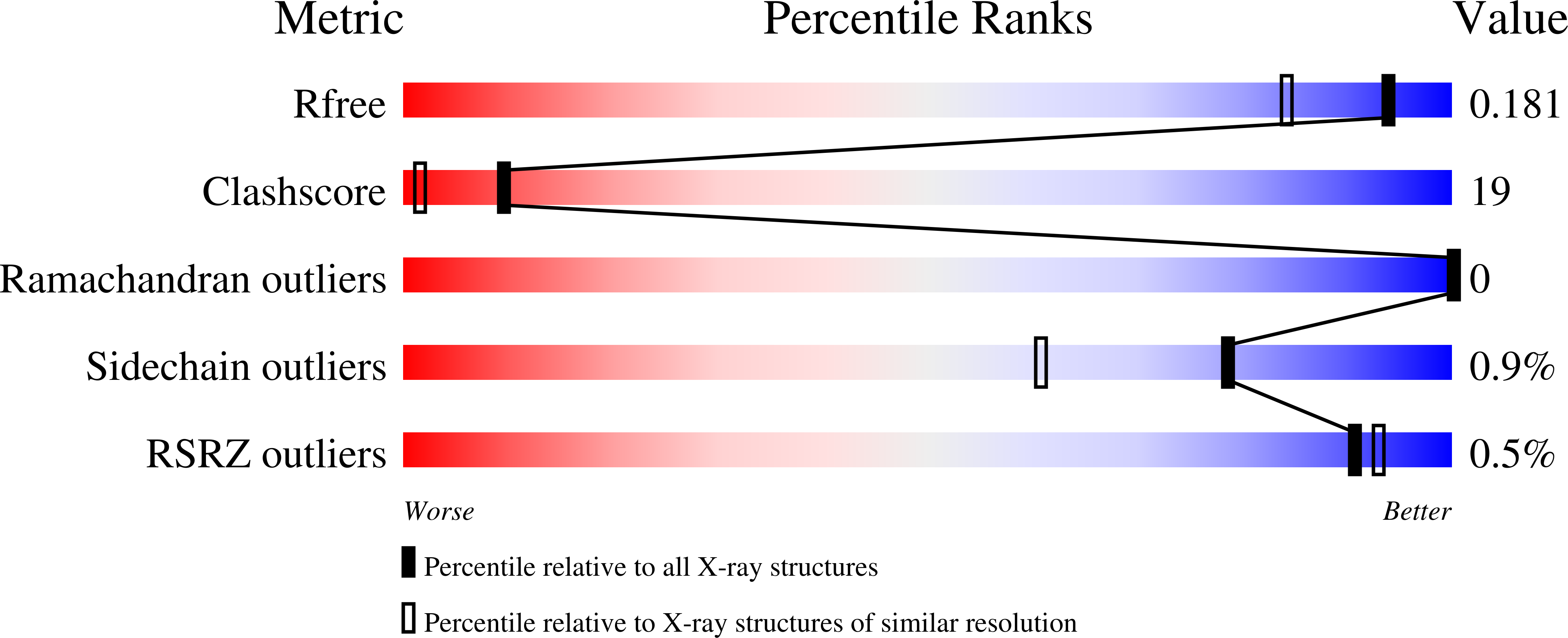
A 1.5 Å resolution X-ray structure of the catalytic module of Caldicellulosiruptor bescii family 3 pectate lyase is reported (PDB entry 3t9g). The resulting structure was refined to an R factor of 0.143 and an R(free) of 0.178. Structural analysis shows that this new structure is very similar to the previously solved structure of a family 3 pectate lyase from Bacillus sp. strain KSM-P15 (PDB entry 1ee6), with a root-mean-square deviation of 0.93 Å and a sequence identity of 53%. This structural similarity is significant considering that C. bescii is a hyperthermophile and Bacillus sp. is a mesophile.
Organizational Affiliation:
BioSciences Center, National Renewable Energy Laboratory, Golden, CO 80401, USA.