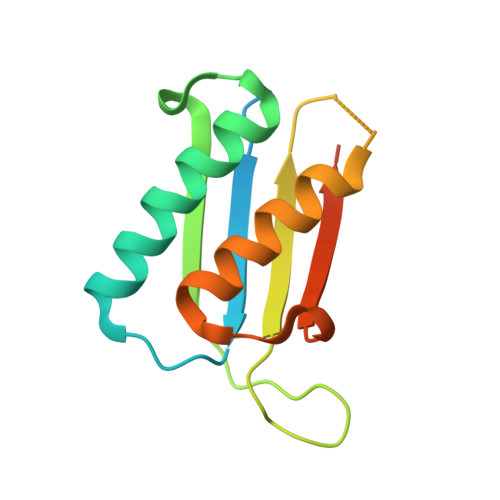
Crystal structure of a macrophage migration inhibitory factor from Giardia lamblia.
Buchko, G.W., Abendroth, J., Robinson, H., Zhang, Y., Hewitt, S.N., Edwards, T.E., Van Voorhis, W.C., Myler, P.J.(2013) J Struct Funct Genomics 14: 47-57
- PubMed: 23709284
- DOI: https://doi.org/10.1007/s10969-013-9155-9
- Primary Citation of Related Structures:
3T5S - PubMed Abstract:
Macrophage migration inhibitory factor (MIF) is a eukaryotic cytokine that affects a broad spectrum of immune responses and its activation/inactivation is associated with numerous diseases. During protozoan infections MIF is not only expressed by the host, but, has also been observed to be expressed by some parasites and released into the host. To better understand the biological role of parasitic MIF proteins, the crystal structure of the MIF protein from Giardia lamblia (Gl-MIF), the etiological agent responsible for giardiasis, has been determined at 2.30 Å resolution. The 114-residue protein adopts an α/β fold consisting of a four-stranded β-sheet with two anti-parallel α-helices packed against a face of the β-sheet. An additional short β-strand aligns anti-parallel to β4 of the β-sheet in the adjacent protein unit to help stabilize a trimer, the biologically relevant unit observed in all solved MIF crystal structures to date, and form a discontinuous β-barrel. The structure of Gl-MIF is compared to the MIF structures from humans (Hs-MIF) and three Plasmodium species (falciparum, berghei, and yoelii). The structure of all five MIF proteins are generally similar with the exception of a channel that runs through the center of each trimer complex. Relative to Hs-MIF, there are differences in solvent accessibility and electrostatic potential distribution in the channel of Gl-MIF and the Plasmodium-MIFs due primarily to two "gate-keeper" residues in the parasitic MIFs. For the Plasmodium MIFs the gate-keeper residues are at positions 44 (Y --> R) and 100 (V --> D) and for Gl-MIF it is at position 100 (V --> R). If these gate-keeper residues have a biological function and contribute to the progression of parasitemia they may also form the basis for structure-based drug design targeting parasitic MIF proteins.
Organizational Affiliation:
Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, USA. garry.buchko@pnnl.gov