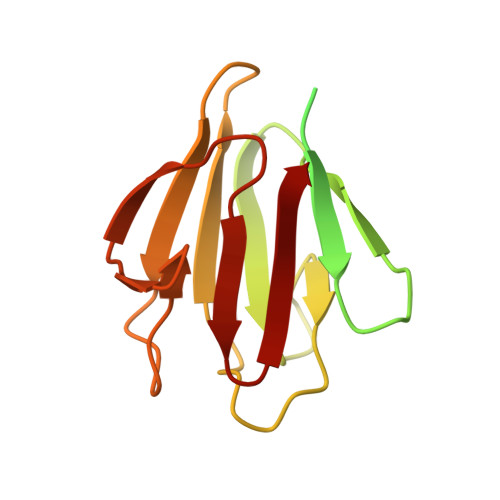
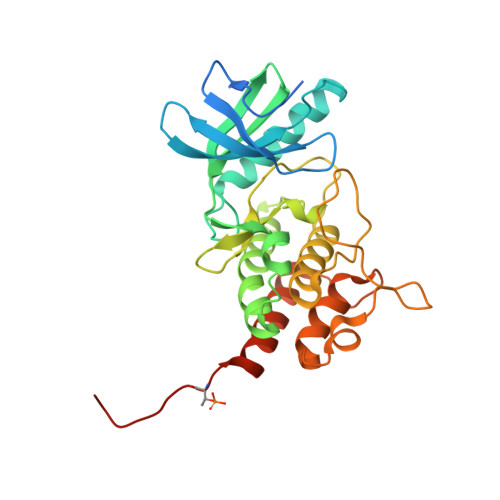
A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria
Gee, C.L., Papavinasasundaram, K.G., Blair, S.R., Baer, C.E., Falick, A.M., King, D.S., Griffin, J.E., Venghatakrishnan, H., Zukauskas, A., Wei, J.R., Dhiman, R.K., Crick, D.C., Rubin, E.J., Sassetti, C.M., Alber, T.(2012) Sci Signal 5: ra7-ra7
- PubMed: 22275220
- DOI: https://doi.org/10.1126/scisignal.2002525
- Primary Citation of Related Structures:
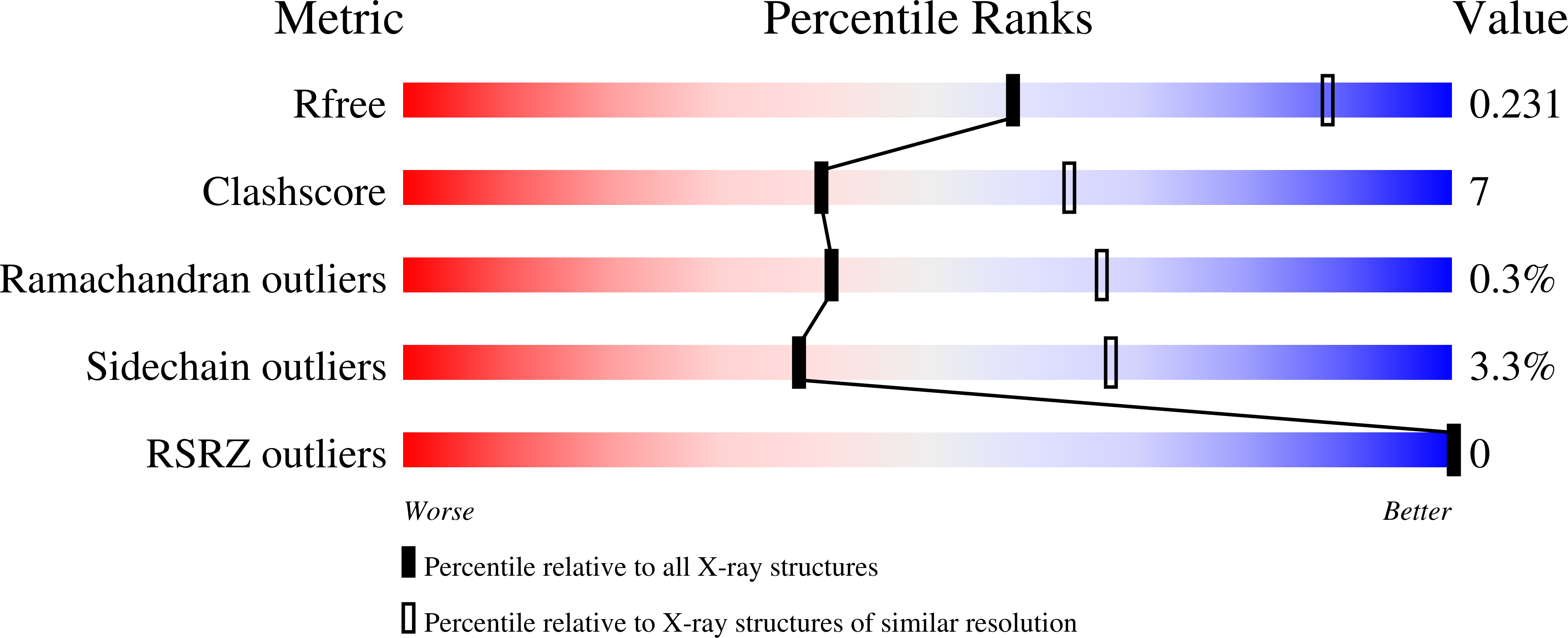
3OTV, 3OUK, 3OUN, 3UQC - PubMed Abstract:
Prokaryotic cell wall biosynthesis is coordinated with cell growth and division, but the mechanisms regulating this dynamic process remain obscure. Here, we describe a phosphorylation-dependent regulatory complex that controls peptidoglycan (PG) biosynthesis in Mycobacterium tuberculosis. We found that PknB, a PG-responsive Ser-Thr protein kinase (STPK), initiates complex assembly by phosphorylating a kinase-like domain in the essential PG biosynthetic protein, MviN. This domain was structurally diverged from active kinases and did not mediate phosphotransfer. Threonine phosphorylation of the pseudokinase domain recruited the FhaA protein through its forkhead-associated (FHA) domain. The crystal structure of this phosphorylated pseudokinase-FHA domain complex revealed the basis of FHA domain recognition, which included unexpected contacts distal to the phosphorylated threonine. Conditional degradation of these proteins in mycobacteria demonstrated that MviN was essential for growth and PG biosynthesis and that FhaA regulated these processes at the cell poles and septum. Controlling this spatially localized PG regulatory complex is only one of several cellular roles ascribed to PknB, suggesting that the capacity to coordinate signaling across multiple processes is an important feature conserved between eukaryotic and prokaryotic STPK networks.
Organizational Affiliation:
Department of Molecular and Cell Biology, QB3 Institute, University of California, Berkeley, Berkeley, CA 94720-3220, USA.