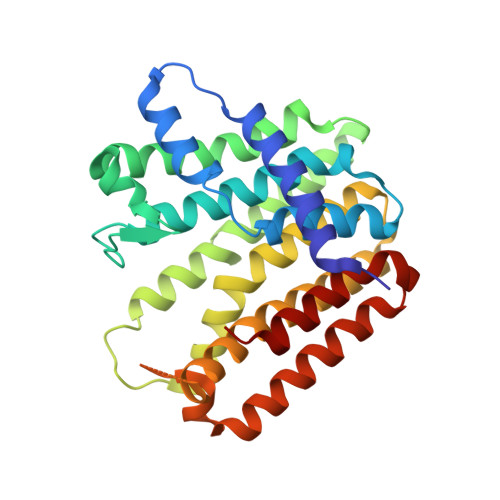
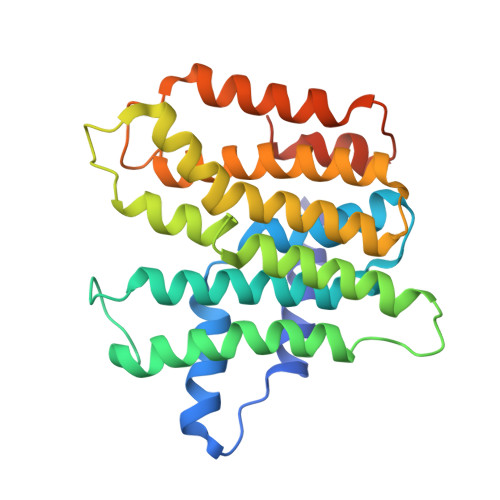
Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions
Hsieh, F.-L., Chang, T.-H., Ko, T.-P., Wang, A.H.-J.(2010) J Mol Biol 404: 859-873
- PubMed: 20965200
- DOI: https://doi.org/10.1016/j.jmb.2010.10.011
- Primary Citation of Related Structures:
3OAB, 3OAC - PubMed Abstract:
Isoprenoids, most of them synthesized by prenyltransferases (PTSs), are a class of important biologically active compounds with diverse functions. The mint geranyl pyrophosphate synthase (GPPS) is a heterotetramer composed of two LSU·SSU (large/small subunit) dimers. In addition to C(10)-GPP, the enzyme also produces geranylgeranyl pyrophosphate (C(20)-GGPP) in vitro, probably because of the conserved active-site structures between the LSU of mint GPPS and the homodimeric GGPP synthase from mustard. By contrast, the SSU lacks the conserved aspartate-rich motifs for catalysis. A major active-site cavity loop in the LSU and other trans-type PTSs is replaced by the regulatory R-loop in the SSU. Only C(10)-GPP, but not C(20)-GGPP, was produced when intersubunit interactions of the R-loop were disrupted by either deletion or multiple point mutations. The structure of the deletion mutant, determined in two different crystal forms, shows an intact (LSU·SSU)(2) heterotetramer, as previously observed in the wild-type enzyme. The active-site of LSU remains largely unaltered, except being slightly more open to the bulk solvent. The R-loop of SSU acts by regulating the product release from LSU, just as does its equivalent loop in a homodimeric PTS, which prevents the early reaction intermediates from escaping the active site of the other subunit. In this way, the product-retaining function of R-loop provides a more stringent control for chain-length determination, complementary to the well-established molecular ruler mechanism. We conclude that the R-loop may be used not only to conserve the GPPS activity but also to produce portions of C(20)-GGPP in mint.
Organizational Affiliation:
Institute of Biological Chemistry, Academia Sinica, Taipei 115, Taiwan.