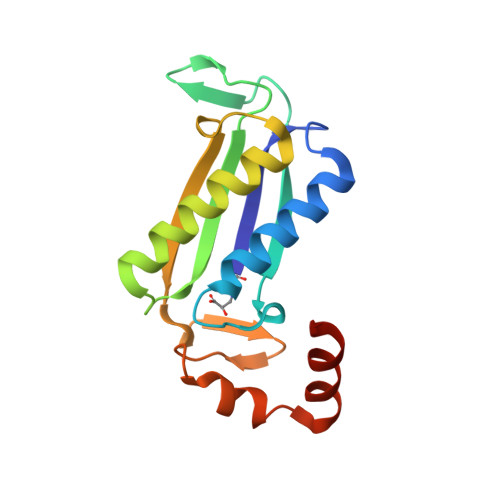
Crystal Structures of the Native and Inactivated Cg10062, a cis-3-Chloroacrylic Acid Dehalogenase from Corynebacterium glutamicum: Implications for the Evolution of cis-3-Chloroacrylic Acid Dehalogenase Activity in the Tautomerase Superfamily
Guo, Y., Robertson, B.A., J Poelarends, G., Thunnissen, A.-M.W.H., Hackert, M.L., Whitman, C.P., Dijkstra, B.W.To be published.