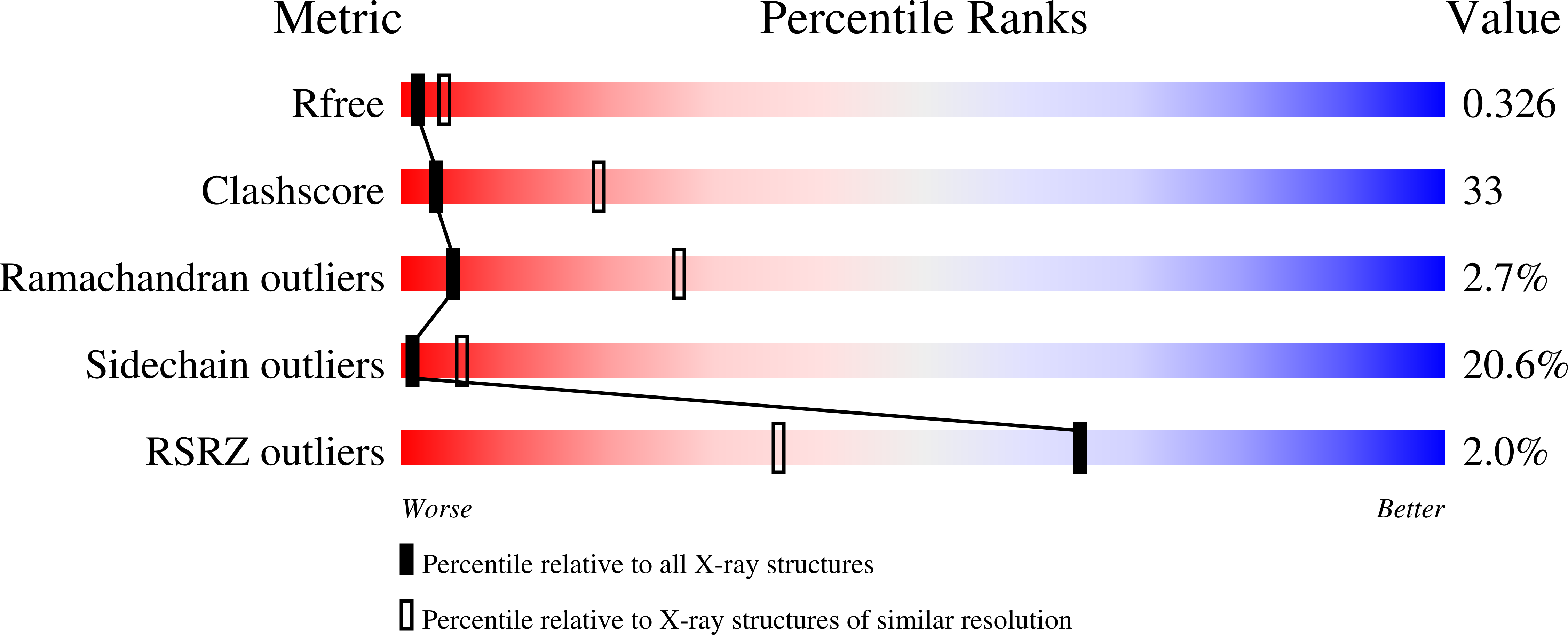
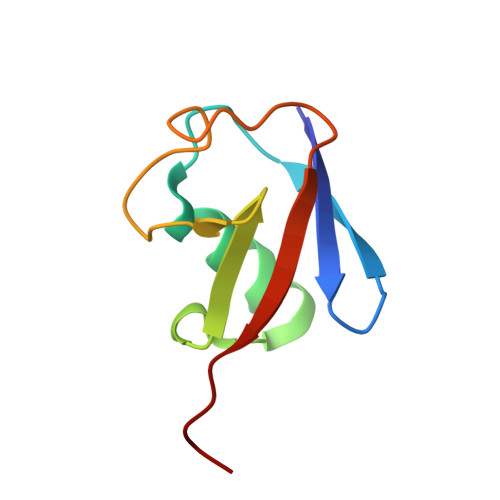
Crystallographic analysis of metal-ion binding to human ubiquitin.
Arnesano, F., Belviso, B.D., Caliandro, R., Falini, G., Fermani, S., Natile, G., Siliqi, D.(2011) Chemistry 17: 1569-1578
- PubMed: 21268159
- DOI: https://doi.org/10.1002/chem.201001617
- Primary Citation of Related Structures:
3N30, 3N32 - PubMed Abstract:
The metal-binding ability of human ubiquitin (hUb) towards a selection of biologically relevant metal ions and complexes has been probed. Different techniques have been used to obtain crystals suitable for crystallographic analysis. In the first type of experiments, crystals of hUb have been soaked in solutions containing copper(II) acetate and two metallodrugs, Zeise salt (K[PtCl(3)(η(2)-C(2)H(4))]·H(2)O) and cisplatin (cis-[PtCl(2)(NH(3))(2)]). The Zeise salt is used in a test for hepatitis, whereas cisplatin is one of the most powerful anticancer drugs in clinical use. The Zeise salt readily reacts with hUb crystals to afford an adduct with three platinum residues per protein molecule, Pt(3)-hUb. In contrast, copper(II) acetate and cisplatin were found to be unreactive for contact times up to one hour and to cause degradation of the hUb crystals for longer times. In the second type of experiments, hUb was cocrystallized with a solution of copper(II) or zinc(II) acetate or cisplatin. Zinc(II) acetate gives, at low metal-to-protein molar ratios (8:1), crystals containing one metal ion per three molecules of protein, Zn-hUb(3) (already reported in previous work), whereas at high metal-to-protein ratios (70:1) gives crystals containing three Zn(II) ions per protein molecule, Zn(3)-hUb. In contrast, once again, copper(II) acetate and cisplatin, even at low metal-to-protein ratios, do not give crystalline material. In the soaking experiment, the Zeise anion leads to simultaneous platination of His68, Met1, and Lys6. Present and previous results of cocrystallization experiments performed with Zn(II) and other Group 12 metal ions allow a comprehensive understanding of the metal-ion binding properties of hUb with His68 as the main anchoring site, followed by Met1 and carboxylic groups of Glu16, Glu18, Glu64, Asp21, and Asp32, to be reached. In the case of platinum, Lys6 can also be a binding site. The amount of bound metal ion, with respect to that of the protein, appears to be a relevant parameter influencing crystal packing.
Organizational Affiliation:
Dipartimento Farmaco-Chimico, University of Bari A. Moro, via E. Orabona 4, 70125 Bari, Italy.