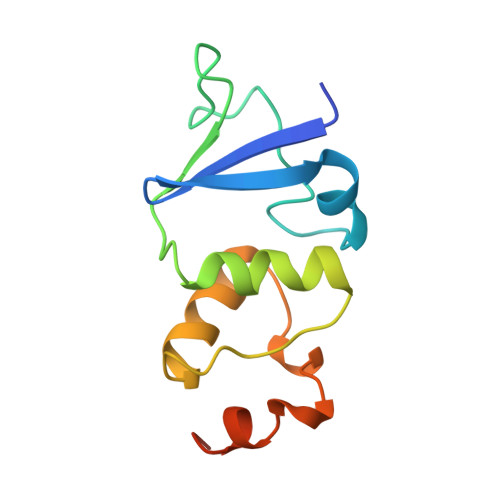
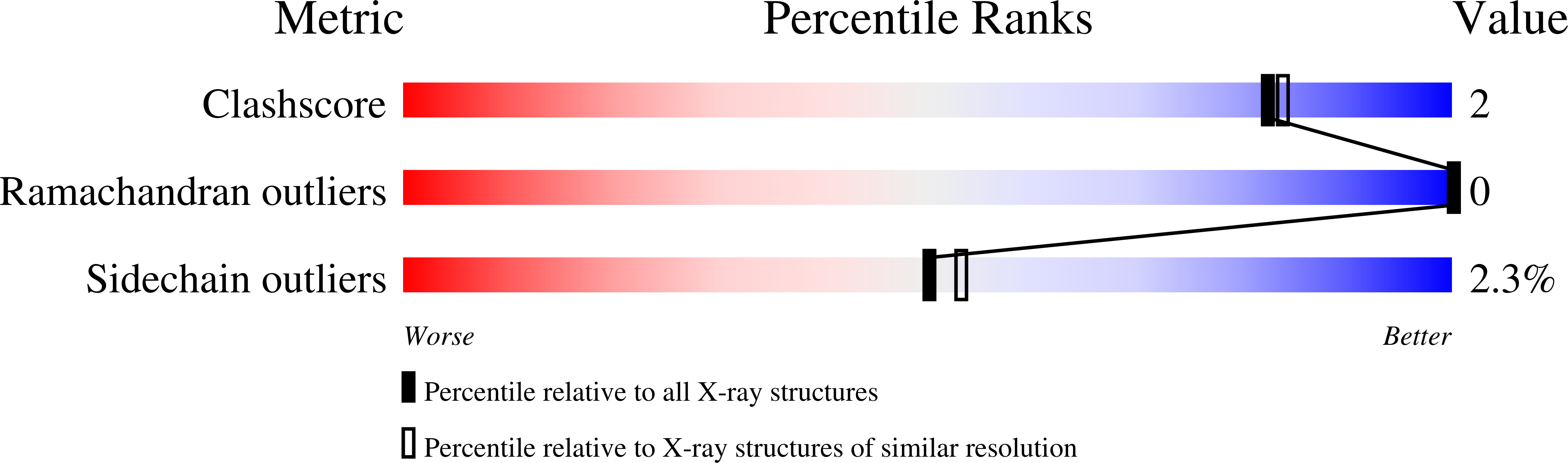Zn2+-binding and molecular determinants of tetramerization in voltage-gated K+ channels.
Bixby, K.A., Nanao, M.H., Shen, N.V., Kreusch, A., Bellamy, H., Pfaffinger, P.J., Choe, S.(1999) Nat Struct Biol 6: 38-43
- PubMed: 9886290
- DOI: https://doi.org/10.1038/4911
- Primary Citation of Related Structures:
1T1D, 3KVT - PubMed Abstract:
The N-terminal, cytoplasmic tetramerization domain (T1) of voltage-gated K+ channels encodes molecular determinants for subfamily-specific assembly of alpha-subunits into functional tetrameric channels. Crystal structures of T1 tetramers from Shaw and Shaker subfamilies reveal a common four-layered scaffolding. Within layer 4, on the hypothetical membrane-facing side of the tetramer, the Shaw T1 tetramer contains four zinc ions; each is coordinated by a histidine and two cysteines from one monomer and by one cysteine from an adjacent monomer. The amino acids involved in coordinating the Zn2+ ion occur in a HX5CX20CC sequence motif that is highly conserved among all Shab, Shaw and Shal subfamily members, but is not found in Shaker subfamily members. We demonstrate by coimmunoprecipitation that a few characteristic residues in the subunit interface are crucial for subfamily-specific tetramerization of the T1 domains.
Organizational Affiliation:
The Salk Institute, Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla 92037, USA.