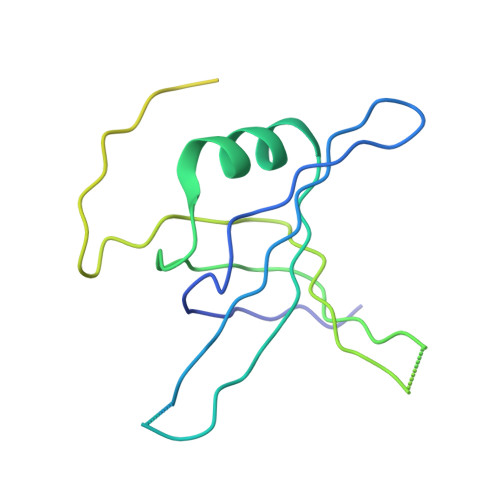
Structure of the single-stranded DNA-binding protein from Streptomyces coelicolor.
Stefanic, Z., Vujaklija, D., Luic, M.(2009) Acta Crystallogr D Biol Crystallogr 65: 974-979
- PubMed: 19690375
- DOI: https://doi.org/10.1107/S0907444909023634
- Primary Citation of Related Structures:
3EIV - PubMed Abstract:
The crystal structure of the single-stranded DNA-binding protein (SSB) from Streptomyces coelicolor, a filamentous soil bacterium with a complex life cycle and a linear chromosome, has been solved and refined at 2.1 A resolution. The three-dimensional structure shows a common conserved central OB-fold that is found in all structurally determined SSB proteins. However, it shows variations in quaternary structure that have previously only been found in mycobacterial SSBs. The strand involved in the clamp mechanism characteristic of this type of quaternary structure leads to higher stability of the homotetramer. To the best of our knowledge, this is the first X-ray structure of an SSB protein from a member of the genus Streptomyces and it was predicted to be the most stable of the structurally characterized bacterial or human mitochondrial SSBs.
Organizational Affiliation:
Rudjer Bosković Institute, Bijenicka cesta 54, Zagreb, Croatia.