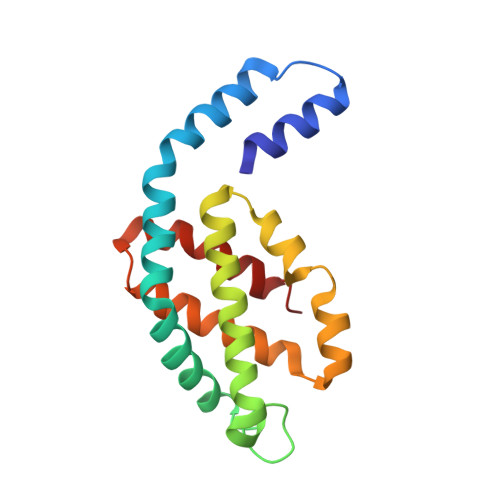
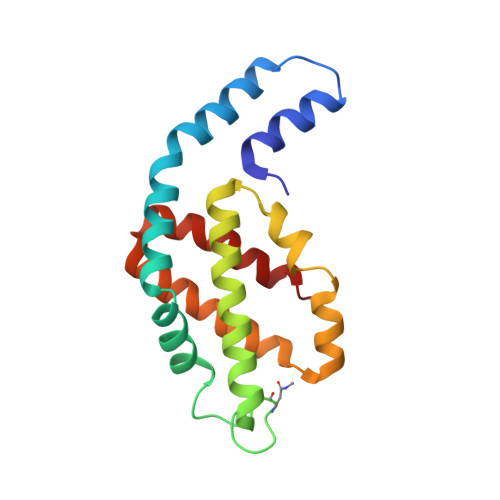
Allophycocyanin Trimer Stability and Functionality Are Primarily Due to Polar Enhanced Hydrophobicity of the Phycocyanobilin Binding Pocket
McGregor, A., Klartag, M., David, L., Adir, N.(2008) J Mol Biol 384: 406-421
- PubMed: 18823993
- DOI: https://doi.org/10.1016/j.jmb.2008.09.018
- Primary Citation of Related Structures:
3DBJ - PubMed Abstract:
Allophycocyanin (APC) is the primary pigment-protein component of the cores of the phycobilisome antenna complex. In addition to an extremely high degree of amino acid sequence conservation, the overall structures of APC from both mesophilic and thermophilic species are almost identical at all levels of assembly, yet APC from thermophilic organisms should have structural attributes that prevent thermally induced denaturation. We determined the structure of APC from the thermophilic cyanobacterium Thermosynechococcus vulcanus to 2.9 A, reaffirming the conservation of structural similarity with APC from mesophiles. We provide spectroscopic evidence that T. vulcanus APC is indeed more stable at elevated temperatures in vitro, when compared with the APC from mesophilic species. APC thermal and chemical stability levels are further enhanced when monitored in the presence of high concentrations of buffered phosphate, which increases the strength of hydrophobic interactions, and may mimic the effect of cytosolic crowding. Absorption spectroscopy, size-exclusion HPLC, and native gel electrophoresis also show that the thermally or chemically induced changes in the APC absorption spectra that result in the loss of the prominent 652-nm band in trimeric APC are not a result of physical monomerization. We propose that the bathochromic shift that occurs in APC upon trimerization is due to the coupling of the hydrophobicity of the alpha84 phycocyanobilin cofactor environment created by a deep cleft formed by the beta subunit with highly charged flanking regions. This arrangement also provides the additional stability required by thermophiles at elevated temperatures. The chemical environment that induces the bathochromic shift in APC trimers is different from the source of shifts in the absorption of monomers of the terminal energy acceptors APC(B) and L(CM), as visualized by the building of molecular models.
Organizational Affiliation:
Schulich Faculty of Chemistry, Technion-Israel Institute of Technology, Technion City, Haifa 32000, Israel.