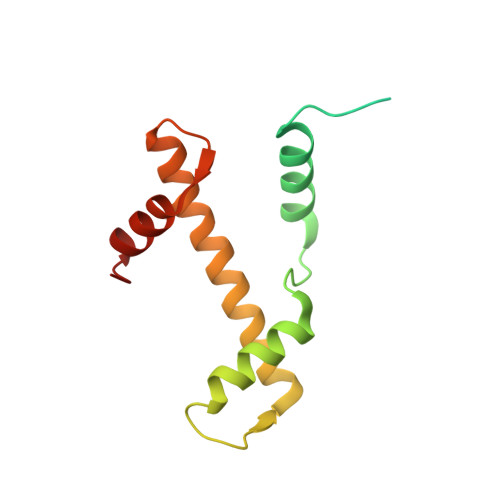
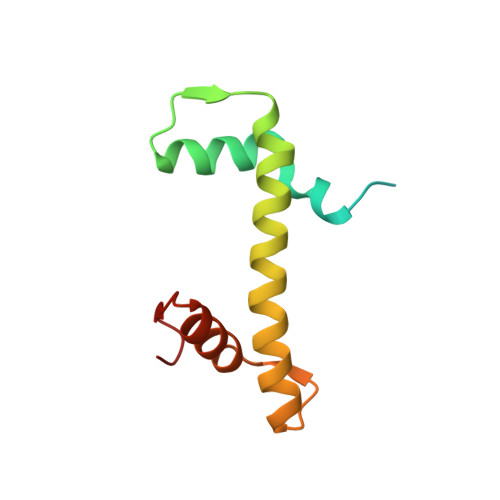
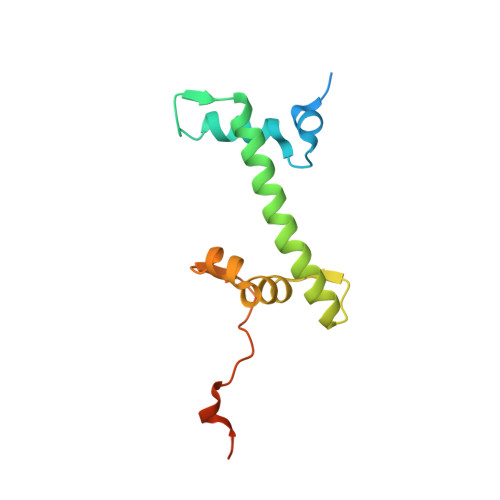
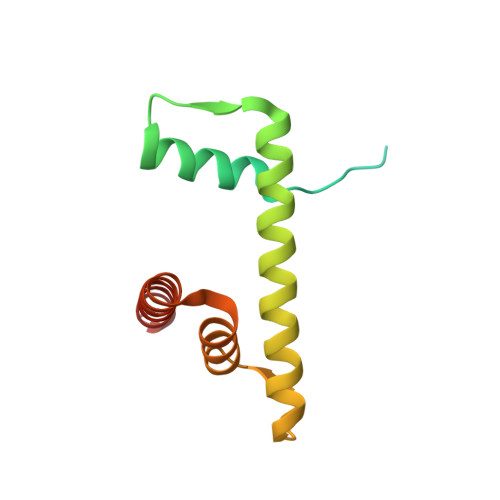
Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Tachiwana, H., Kagawa, W., Osakabe, A., Kawaguchi, K., Shiga, T., Hayashi-Takanaka, Y., Kimura, H., Kurumizaka, H.(2010) Proc Natl Acad Sci U S A 107: 10454-10459
- PubMed: 20498094
- DOI: https://doi.org/10.1073/pnas.1003064107
- Primary Citation of Related Structures:
3A6N, 3AFA - PubMed Abstract:
A histone H3 variant, H3T, is highly expressed in the testis, suggesting that it may play an important role in the chromatin reorganization required for meiosis and/or spermatogenesis. In the present study, we found that the nucleosome containing human H3T is significantly unstable both in vitro and in vivo, as compared to the conventional nucleosome containing H3.1. The crystal structure of the H3T nucleosome revealed structural differences in the H3T regions on both ends of the central alpha2 helix, as compared to those of H3.1. The H3T-specific residues (Met71 and Val111) are the source of the structural differences observed between H3T and H3.1. A mutational analysis revealed that these residues are responsible for the reduced stability of the H3T-containing nucleosome. These physical and structural properties of the H3T-containing nucleosome may provide the basis of chromatin reorganization during spermatogenesis.
Organizational Affiliation:
Laboratory of Structural Biology, Graduate School of Advanced Science and Engineering, Waseda University, Shinjuku-ku, Tokyo 162-8480, Japan.