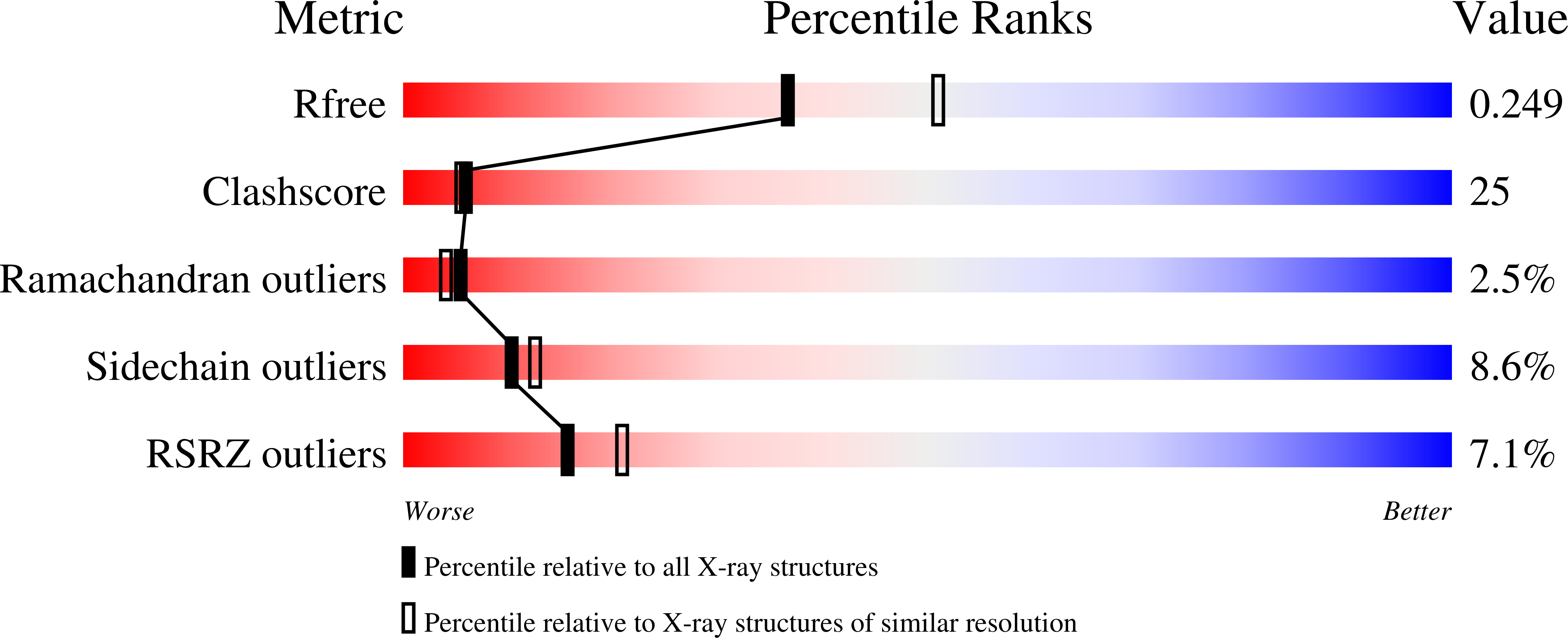
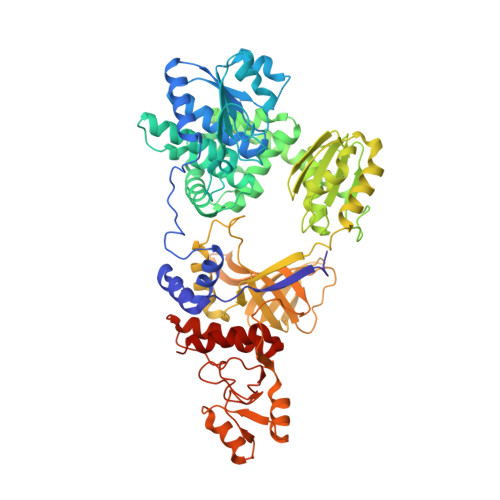
Structure of RecJ exonuclease defines its specificity for single-stranded DNA
Wakamatsu, T., Kitamura, Y., Kotera, Y., Nakagawa, N., Kuramitsu, S., Masui, R.(2010) J Biol Chem 285: 9762-9769
- PubMed: 20129927
- DOI: https://doi.org/10.1074/jbc.M109.096487
- Primary Citation of Related Structures:
2ZXO, 2ZXP, 2ZXR - PubMed Abstract:
RecJ is a single-stranded DNA (ssDNA)-specific 5'-3' exonuclease that plays an important role in DNA repair and recombination. To elucidate how RecJ achieves its high specificity for ssDNA, we determined the entire structures of RecJ both in a ligand-free form and in a complex with Mn(2+) or Mg(2+) by x-ray crystallography. The entire RecJ consists of four domains that form a molecule with an O-like structure. One of two newly identified domains had structural similarities to an oligonucleotide/oligosaccharide-binding (OB) fold. The OB fold domain alone could bind to DNA, indicating that this domain is a novel member of the OB fold superfamily. The truncated RecJ containing only the core domain exhibited much lower affinity for the ssDNA substrate compared with intact RecJ. These results support the hypothesis that these structural features allow specific binding of RecJ to ssDNA. In addition, the structure of the RecJ-Mn(2+) complex suggests that the hydrolysis reaction catalyzed by RecJ proceeds through a two-metal ion mechanism.
Organizational Affiliation:
Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka 565-0871.