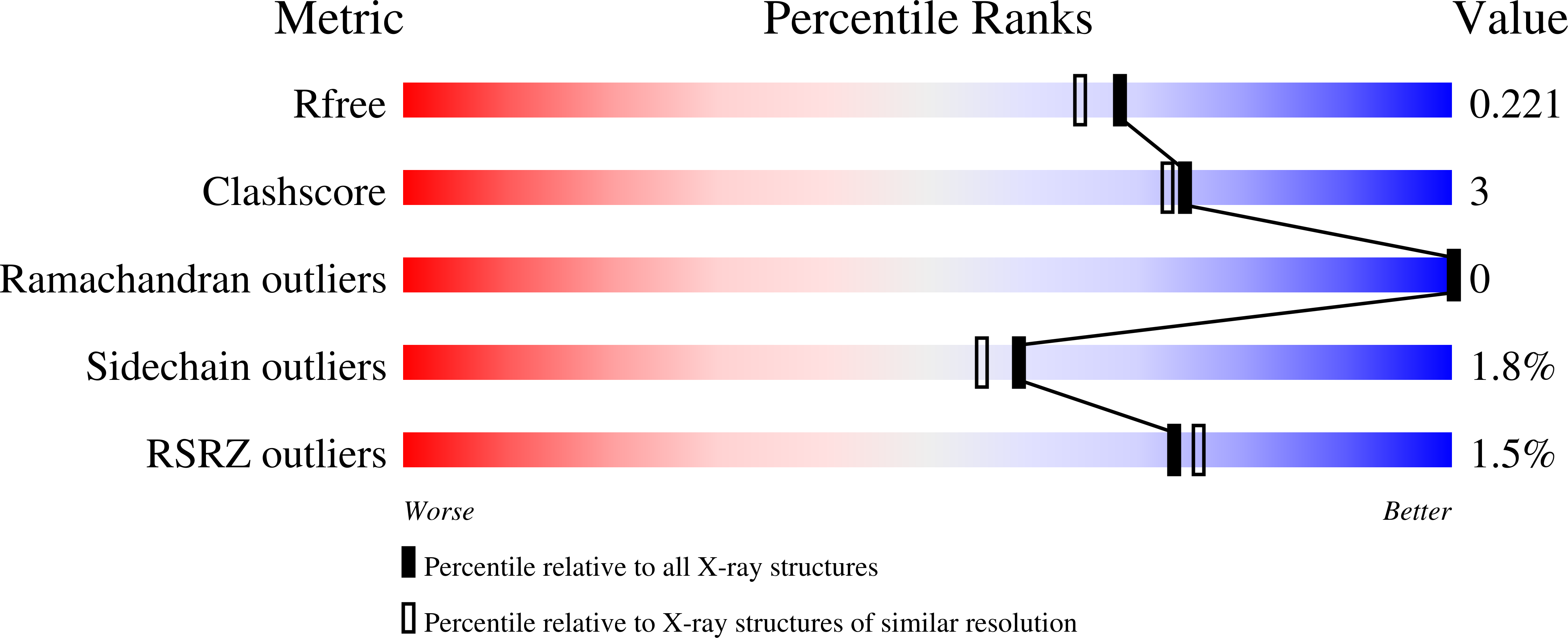
Sugar-complex structures of the C-half domain of the galactose-binding lectin EW29 from the earthworm Lumbricus terrestris
Suzuki, R., Kuno, A., Hasegawa, T., Hirabayashi, J., Kasai, K., Momma, M., Fujimoto, Z.(2009) Acta Crystallogr D Biol Crystallogr 65: 49-57
- PubMed: 19153466
- DOI: https://doi.org/10.1107/S0907444908037451
- Primary Citation of Related Structures:
2ZQN, 2ZQO - PubMed Abstract:
R-type lectins are one of the most prominent types of lectin; they exist ubiquitously in nature and mainly bind to the galactose unit of sugar chains. The galactose-binding lectin EW29 from the earthworm Lumbricus terrestris belongs to the R-type lectin family as represented by the plant lectin ricin. It shows haemagglutination activity and is composed of a single peptide chain that includes two homologous domains: N-terminal and C-terminal domains. A truncated mutant of EW29 comprising the C-terminal domain (rC-half) has haemagglutination activity by itself. In order to clarify how rC-half recognizes ligands and shows haemagglutination activity, X-ray crystal structures of rC-half in complex with D-lactose and N-acetyl-D-galactosamine have been determined. The structure of rC-half is similar to that of the ricin B chain and consists of a beta-trefoil fold; the fold is further divided into three similar subdomains referred to as subdomains alpha, beta and gamma, which are gathered around the pseudo-threefold axis. The structures of sugar complexes demonstrated that subdomains alpha and gamma of rC-half bind terminal galactosyl and N-acetylgalactosaminyl glycans. The sugar-binding properties are common to both ligands in both subdomains and are quite similar to those of ricin B chain-lactose complexes. These results indicate that the C-terminal domain of EW29 uses these two galactose-binding sites for its function as a single-domain-type haemagglutinin.
Organizational Affiliation:
Protein Research Unit, National Institute of Agrobiological Sciences, Tsukuba 305-8602, Japan.