Pan-Modular Structure of Microneme Protein Sml-2 from Parasite Sarcocystis Muris at 1.95 A Resolution and its Complex with 1-Thio-Beta-D-Galactose.
Mueller, J.J., Weiss, M.S., Heinemann, U.(2011) Acta Crystallogr D Biol Crystallogr D67: 936
- PubMed: 22101820
- DOI: https://doi.org/10.1107/S0907444911037796
- Primary Citation of Related Structures:
2YIL, 2YIO, 2YIP - PubMed Abstract:
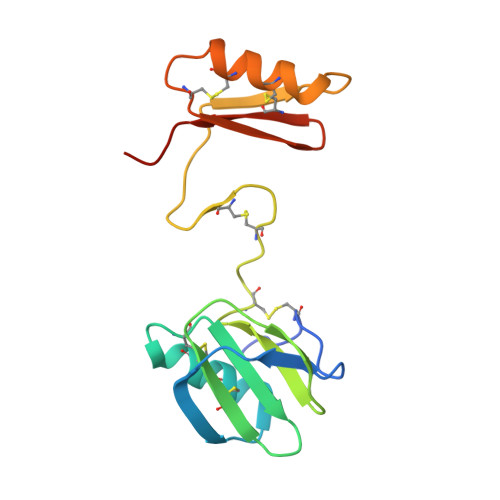
The microneme protein SML-2 is a member of a small family of galactose-specific lectins that play a role during host-cell invasion by the apicomplexan parasite Sarcocystis muris. The structures of apo SML-2 and the 1-thio-β-D-galactose-SML-2 complex were determined at 1.95 and 2.1 Å resolution, respectively, by sulfur-SAD phasing. Highly elongated dimers are formed by PAN-domain tandems in the protomer, bearing the galactose-binding cavities at the distal apple-like domains. The detailed structure of the binding site in SML-2 explains the high specificity of galactose-endgroup binding and the broader specificity of the related Toxoplasma gondii protein TgMIC4 towards galactose and glucose. A large buried surface of highly hydrophobic character and 24 intersubunit hydrogen bonds stabilize the dimers and half of the 12 disulfides per dimer are shielded from the solvent by the polypeptide chain, thereby enhancing the resistance of the parasite protein towards unfolding and proteolysis that allows it to survive within the intestinal tracts of the intermediate and final hosts.
Organizational Affiliation:
Kristallographie, Max-Delbrück-Centrum für Molekulare Medizin, Robert-Rössle-Strasse 10, 13125 Berlin, Germany.