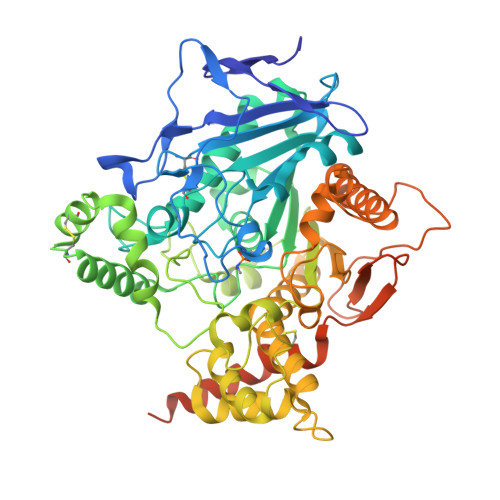
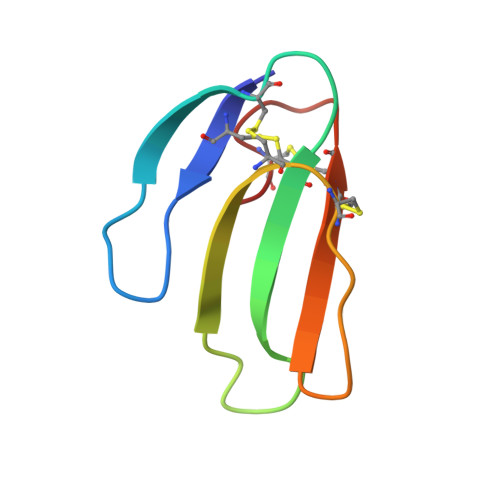
Structural evidence that human acetylcholinesterase inhibited by tabun ages through O-dealkylation.
Carletti, E., Colletier, J.P., Dupeux, F., Trovaslet, M., Masson, P., Nachon, F.(2010) J Med Chem 53: 4002-4008
- PubMed: 20408548
- DOI: https://doi.org/10.1021/jm901853b
- Primary Citation of Related Structures:
2X8B - PubMed Abstract:
Tabun is a warfare agent that inhibits human acetylcholinesterase (hAChE) by rapid phosphylation of the catalytic serine. A time-dependent reaction occurs on the tabun adduct, leading to an "aged" enzyme, resistant to oxime reactivators. The aging reaction may proceed via either dealkylation or deamidation, depending on the stereochemistry of the phosphoramidyl adduct. We solved the X-ray structure of aged tabun-hAChE complexed with fasciculin II, and we show that aging proceeds through O-dealkylation, in agreement with the aging mechanism that we determined for tabun-inhibited human butyrylcholinesterase and mouse acetylcholinesterase. Noteworthy, aging and binding of fasciculin II lead to an improved thermostability, resulting from additional stabilizing interactions between the two subdomains that face each other across the active site gorge. This first structure of hAChE inhibited by a nerve agent provides structural insight into the inhibition and aging mechanisms and a structural template for the design of molecules capable of reactivating aged hAChE.
Organizational Affiliation:
Laboratoire de Biophysique Moléculaire, Institut de Biologie Structurale (CEA-CNRS-UJF), Grenoble, France.