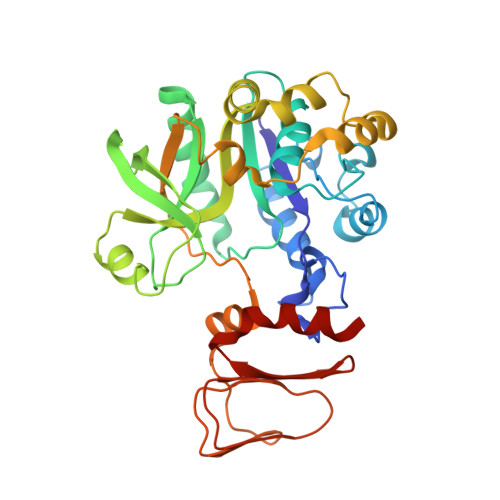
Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
Pelissier, M.C., Lesley, S., Kuhn, P., Bourne, Y.(2010) J Biol Chem 285: 27468
- PubMed: 20573954
- DOI: https://doi.org/10.1074/jbc.M109.095182
- Primary Citation of Related Structures:
2X5S, 2X5Z, 2X60, 2X65 - PubMed Abstract:
GMP catalyzes the formation of GDP-Man, a fundamental precursor for protein glycosylation and bacterial cell wall and capsular polysaccharide biosynthesis. Crystal structures of GMP from the thermophilic bacterium Thermotoga maritima in the apo form, in complex with the substrates mannose-1-phosphate or GTP and bound with the end product GDP-Man in the presence of the essential divalent cation Mg(2+), were solved in the 2.1-2.8 A resolution range. The T. maritima GMP molecule is organized in two separate domains: a N-terminal Rossman fold-like domain and a C-terminal left-handed beta-helix domain. Two molecules associate into a dimer through a tail-to-tail arrangement of the C-terminal domains. Comparative analysis of the structures along with characterization of enzymatic parameters reveals the bases of substrate specificity of this class of sugar nucleotidyltransferases. In particular, substrate and product binding are associated with significant changes in the conformation of loop regions lining the active center and in the relative orientation of the two domains. Involvement of both the N- and C-terminal domains, coupled to the catalytic role of a bivalent metal ion, highlights the catalytic features of bacterial GMPs compared with other members of the pyrophosphorylase superfamily.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, UMR-6098, CNRS, Université Aix-Marseille, F-13288 Marseille, France.