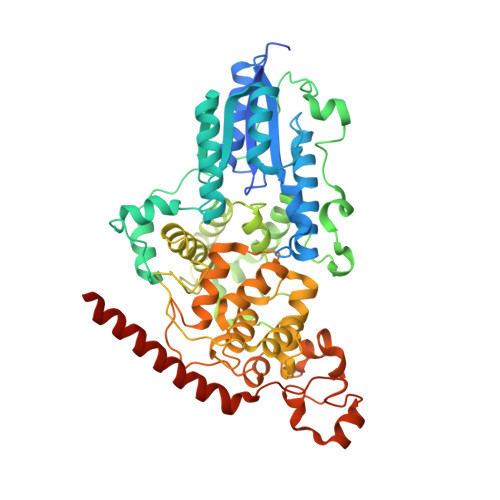
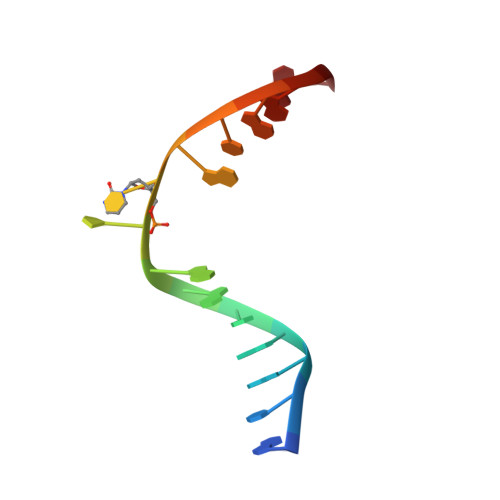
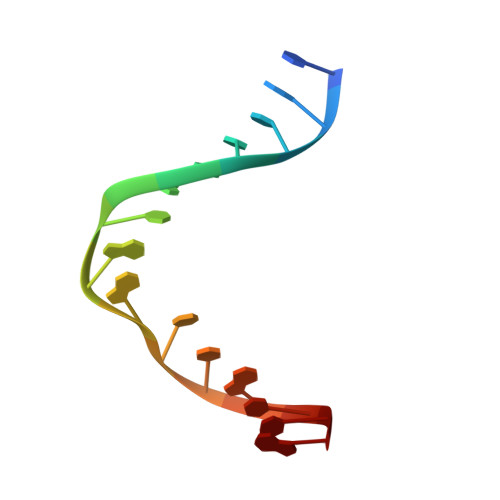
Crystal Structure of the T(6-4)C Lesion in Complex with a (6-4) DNA Photolyase and Repair of Uv- Induced (6-4) and Dewar Photolesions.
Glas, A.F., Schneider, S., Maul, M.J., Hennecke, U., Carell, T.(2009) Chemistry 15: 10387
- PubMed: 19722240
- DOI: https://doi.org/10.1002/chem.200901004
- Primary Citation of Related Structures:
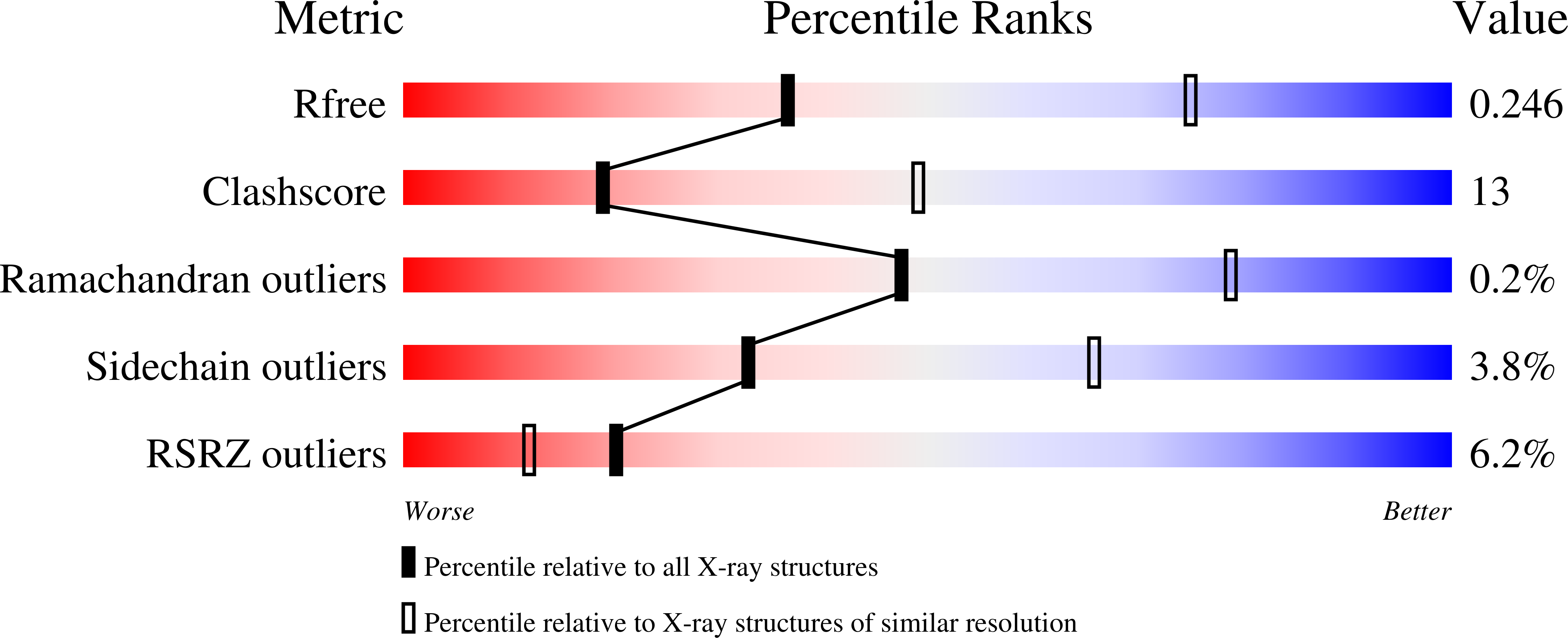
2WB2 - PubMed Abstract:
UV-light irradiation induces the formation of highly mutagenic lesions in DNA, such as cis-syn cyclobutane pyrimidine dimers (CPD photoproducts), pyrimidine(6-4)pyrimidone photoproducts ((6-4) photoproducts) and their Dewar valence isomers ((Dew) photoproducts). Here we describe the synthesis of defined DNA strands containing these lesions by direct irradiation. We show that all lesions are efficiently repaired except for the T(Dew)T lesion, which cannot be cleaved by the repair enzyme under our conditions. A crystal structure of a T(6-4)C lesion containing DNA duplex in complex with the (6-4) photolyase from Drosophila melanogaster provides insight into the molecular recognition event of a cytosine derived photolesion for the first time. In light of the previously postulated repair mechanism, which involves rearrangement of the (6-4) lesions into strained four-membered ring repair intermediates, it is surprising that the not rearranged T(6-4)C lesion is observed in the active site. The structure, therefore, provides additional support for the newly postulated repair mechanism that avoids this rearrangement step and argues for a direct electron injection into the lesion as the first step of the repair reaction performed by (6-4) DNA photolyases.
Organizational Affiliation:
Department for Chemistry and Biochemistry, Ludwig-Maximilians University, Butenandtstr. 5-13, 81377 Munich, Germany.