The Hemoglobins of the Trematodes Fasciola Hepatica and Paramphistomum Epiclitum: A Molecular Biological, Physico-Chemical, Kinetic, and Vaccination Study.
Dewilde, S., Ioanitescu, A.I., Kiger, L., Gilany, K., Marden, M.C., Van Doorslaer, S., Vercruysse, J., Pesce, A., Nardini, M., Bolognesi, M., Moens, L.(2008) Protein Sci 17: 1653
- PubMed: 18621914
- DOI: https://doi.org/10.1110/ps.036558.108
- Primary Citation of Related Structures:
2VYW - PubMed Abstract:
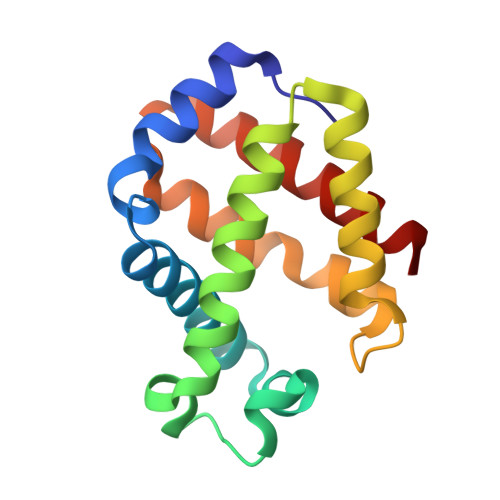
The trematode Fasciola hepatica (Fa.he.) is a common parasite of human and livestock. The hemoglobin (Hb) of Fa.he., a potential immunogen, was chosen for characterization in the search for an effective vaccine. Characterization of trematode Hbs show that they are intracellular single-domain globins with the following remarkable features: (1) Fa.he. expresses two Hb isoforms that differ at two amino acid sites (F1: 119Y/123Q; F2: 119F/123L). Both isoforms are monoacetylated at their N-termini; (2) the genes coding for Fa.he. and Paramphistomum epiclitum (Pa.ep.) Hbs are interrupted by two introns at the conserved positions B12.2 and G7.0.; (3) UV/VIS and resonance Raman spectroscopy identify the recombinant Fa.he. HbF2 as a pentacoordinated high-spin ferrous Hb; (4) electron paramagnetic resonance spectroscopy of cyano-met Fa.he. HbF2 proves that the endogenously bound imidazole has no imidazolate character; (5) the major structural determinants of the globin fold are present, they contain a TyrB10/TyrE7 residue pair on the distal side. Although such distal-site pair is a signature for high oxygen affinity, as shown for Pa.ep. Hb, the oxygen-binding rate parameters for Fa.he. Hb are intermediate between those of myoglobin and those of other trematode Hbs; (6) the three-dimensional structure of recombinant Fa.he. HbF2 from this study closely resembles the three-dimensional structure of Pa.ep. determined earlier. The set of distal-site polar interactions observed in Pa.ep. Hb is matched with small but significant structural adjustments; (7) despite the potential immunogenic character of the fluke Hb, vaccination of calves with recombinant Fa.he. HbF2 failed to promote protection against parasitic infection.
Organizational Affiliation:
Department of Biomedical Sciences, University of Antwerp, Universiteitsplein 1, B-2610 Antwerp, Belgium. sylvia.dewilde@ua.ac.be