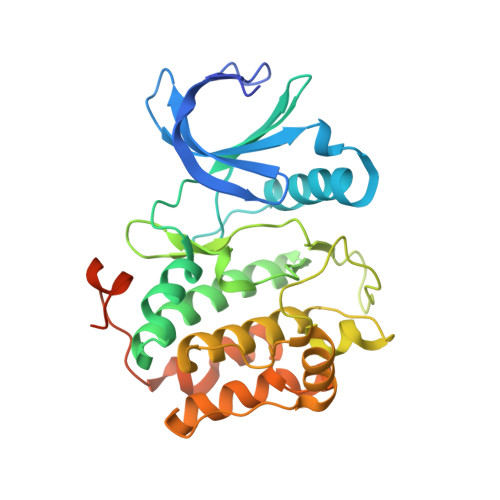
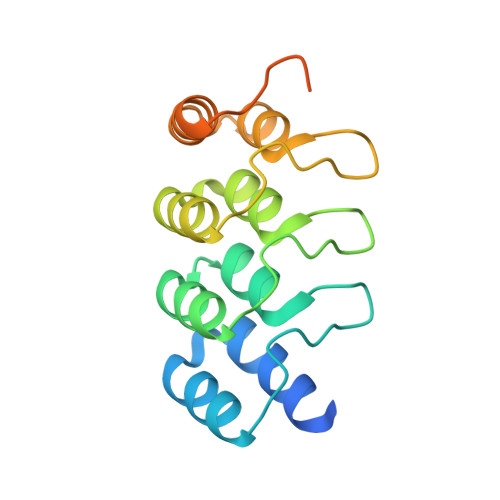
Structure of wild-type Plk-1 kinase domain in complex with a selective DARPin.
Bandeiras, T.M., Hillig, R.C., Matias, P.M., Eberspaecher, U., Fanghanel, J., Thomaz, M., Miranda, S., Crusius, K., Putter, V., Amstutz, P., Gulotti-Georgieva, M., Binz, H.K., Holz, C., Schmitz, A.A., Lang, C., Donner, P., Egner, U., Carrondo, M.A., Muller-Tiemann, B.(2008) Acta Crystallogr D Biol Crystallogr 64: 339-353
- PubMed: 18391401
- DOI: https://doi.org/10.1107/S0907444907068217
- Primary Citation of Related Structures:
2V5Q - PubMed Abstract:
As a key regulator of mitosis, the Ser/Thr protein polo-like kinase-1 (Plk-1) is a well validated drug target in cancer therapy. In order to enable structure-guided drug design, determination of the crystal structure of the kinase domain of Plk-1 was attempted. Using a multi-parallel cloning and expression approach, a set of length variants were identified which could be expressed in large amounts from insect cells and which could be purified to high purity. However, all attempts to crystallize these constructs failed. Crystals were ultimately obtained by generating designed ankyrin-repeat proteins (DARPins) selective for Plk-1 and using them for cocrystallization. Here, the first crystal structure of the kinase domain of wild-type apo Plk-1, in complex with DARPin 3H10, is presented, underlining the power of selective DARPins as crystallization tools. The structure was refined to 2.3 A resolution and shows the active conformation of Plk-1. It broadens the basis for modelling and cocrystallization studies for drug design. The binding epitope of 3H10 is rich in arginine, glutamine and lysine residues, suggesting that the DARPin enabled crystallization by masking a surface patch which is unfavourable for crystal contact formation. Based on the packing observed in the crystal, a truncated DARPin variant was designed which showed improved binding characteristics.
Organizational Affiliation:
Instituto de Biologia Experimental e Tecnológica, Apartado 12, 2780 Oeiras, Portugal.