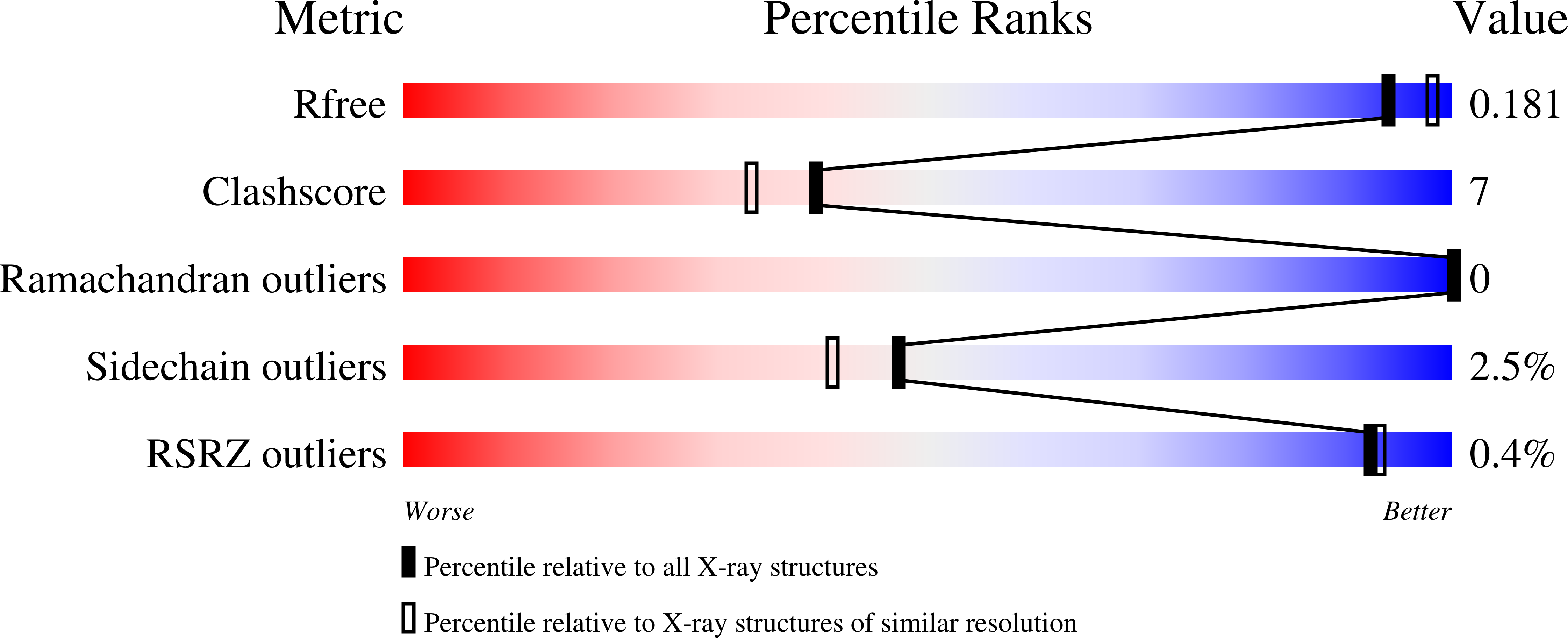
Structural Studies Show that the A178L Mutation in the C-Terminal Hinge of the Catalytic Loop-6 of Triosephosphate Isomerase (Tim) Induces a Closed-Like Conformation in Dimeric and Monomeric Tim.
Alahuhta, M., Casteleijn, M.G., Neubauer, P., Wierenga, R.K.(2008) Acta Crystallogr D Biol Crystallogr 64: 178
- PubMed: 18219118
- DOI: https://doi.org/10.1107/S0907444907059021
- Primary Citation of Related Structures:
2V0T, 2V2C, 2V2D, 2V2H - PubMed Abstract:
The flexible catalytic loop, loop-6, of TIM has evolved to preferably be open in the unliganded state and to preferably be closed in the liganded state. The N-terminal and C-terminal hinges of this loop are important for its opening/closing mechanism. In this study, a small conserved C-terminal hinge residue, Ala178, has been mutated into a residue with a larger side chain, Leu178. This mutation has been made in the dimeric trypanosomal wild-type TIM (wtTIM) and in its mutated catalytically competent monomeric variant (ml1TIM). The variants are referred to as A178L and ml1A178L, respectively. Crystal structures have been determined of unliganded A178L (at 2.2 A), liganded A178L (at 1.89 A), unliganded ml1A178L (at 2.3 A) and liganded ml1A178L (at 1.18 A) using the transition-state analogue 2-phosphoglycolate as a ligand. Structural characterization of the two variants shows that this mutation favours the closed conformation of the C-terminal hinge region, even in the absence of ligand. In the structure of the unliganded A178L variant a range of new loop-6 conformations are observed, including subunits in which the tip of loop-6 is completely disordered. The catalytic efficiency of A178L is lower than that of wtTIM, which correlates with the structural differences between the apo forms of wtTIM and A178L, in particular the more disordered loop-6 in the structure of unliganded A178L. In the liganded structures of A178L and ml1A178L the structural differences induced by the mutation are minimal. Structural characterization of the ml1A178L variant highlights its structural plasticity.
Organizational Affiliation:
Biocenter Oulu and Department of Biochemistry, University of Oulu, PO Box 3000, FIN-90014 University of Oulu, Finland.