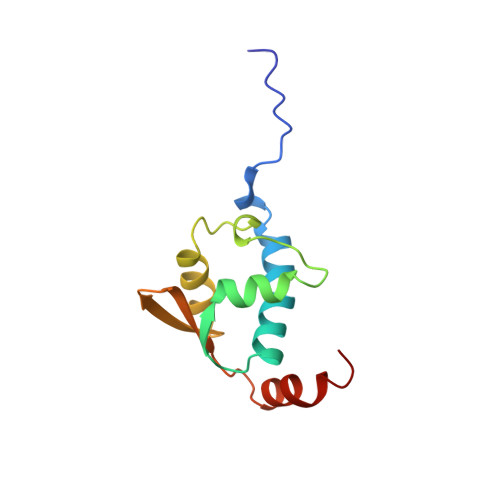
Solution Structure of the Region 51-160 of Human Kin17 Reveals an Atypical Winged Helix Domain
Carlier, L., Le Maire, A., Gondry, M., Guilhaudis, L., Milazzo, I., Davoust, D., Couprie, J., Gilquin, B., Zinn-Justin, S.(2007) Protein Sci 16: 2750
- PubMed: 18029424
- DOI: https://doi.org/10.1110/ps.073079107
- Primary Citation of Related Structures:
2V1N - PubMed Abstract:
Human KIN17 is a 45-kDa eukaryotic DNA- and RNA-binding protein that plays an important role in nuclear metabolism and in particular in the general response to genotoxics. Its amino acids sequence contains a zinc finger motif (residues 28-50) within a 30-kDa N-terminal region conserved from yeast to human, and a 15-kDa C-terminal tandem of SH3-like subdomains (residues 268-393) only found in higher eukaryotes. Here we report the solution structure of the region 51-160 of human KIN17. We show that this fragment folds into a three-alpha-helix bundle packed against a three-stranded beta-sheet. It belongs to the winged helix (WH) family. Structural comparison with analogous WH domains reveals that KIN17 WH module presents an additional and highly conserved 3(10)-helix. Moreover, KIN17 WH helix H3 is not positively charged as in classical DNA-binding WH domains. Thus, human KIN17 region 51-160 might rather be involved in protein-protein interaction through its conserved surface centered on the 3(10)-helix.
Organizational Affiliation:
Equipe de Chimie Organique et Biologie Structurale, IFRMP 23, CNRS UMR 6014, Université de Rouen, 76821 Mont-Saint-Aignan, France.