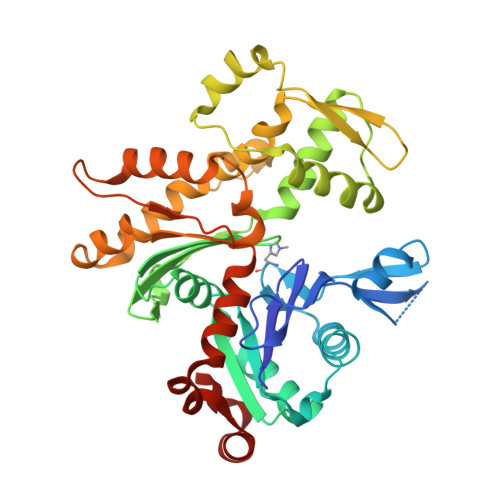
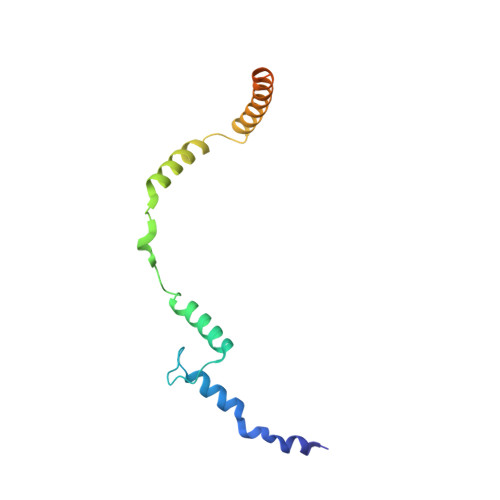
Toxofilin from Toxoplasma gondii forms a ternary complex with an antiparallel actin dimer
Lee, S.H., Hayes, D.B., Rebowski, G., Tardieux, I., Dominguez, R.(2007) Proc Natl Acad Sci U S A 104: 16122-16127
- PubMed: 17911258
- DOI: https://doi.org/10.1073/pnas.0705794104
- Primary Citation of Related Structures:
2Q97 - PubMed Abstract:
Many human pathogens exploit the actin cytoskeleton during infection, including Toxoplasma gondii, an apicomplexan parasite related to Plasmodium, the agent of malaria. One of the most abundantly expressed proteins of T. gondii is toxofilin, a monomeric actin-binding protein (ABP) involved in invasion. Toxofilin is found in rhoptry and presents an N-terminal signal sequence, consistent with its being secreted during invasion. We report the structure of toxofilin amino acids 69-196 in complex with the host mammalian actin. Toxofilin presents an extended conformation and interacts with an antiparallel actin dimer, in which one of the actins is related by crystal symmetry. Consistent with this observation, analytical ultracentrifugation analysis shows that toxofilin binds two actins in solution. Toxofilin folds into five consecutive helices, which form three relatively independent actin-binding sites. Helices 1 and 2 bind the symmetry-related actin molecule and cover its nucleotide-binding cleft. Helices 3-5 bind the other actin and constitute the primary actin-binding region. Helix 3 interacts in the cleft between subdomains 1 and 3, a common binding site for most ABPs. Helices 4 and 5 wrap around actin subdomain 4, and residue Gln-134 of helix 4 makes a hydrogen-bonding contact with the nucleotide in actin, both of which are unique features among ABPs. Toxofilin dramatically inhibits nucleotide exchange on two actin molecules simultaneously. This effect is linked to the formation of the antiparallel actin dimer because a construct lacking helices 1 and 2 binds only one actin and inhibits nucleotide exchange less potently.
Organizational Affiliation:
Department of Physiology, University of Pennsylvania School of Medicine, 3700 Hamilton Walk, Philadelphia, PA 19104-6085, USA.