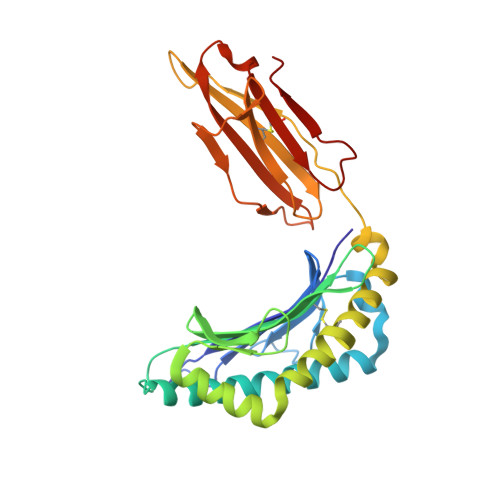
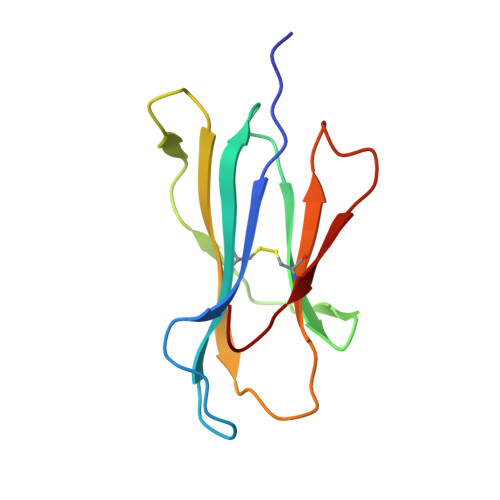
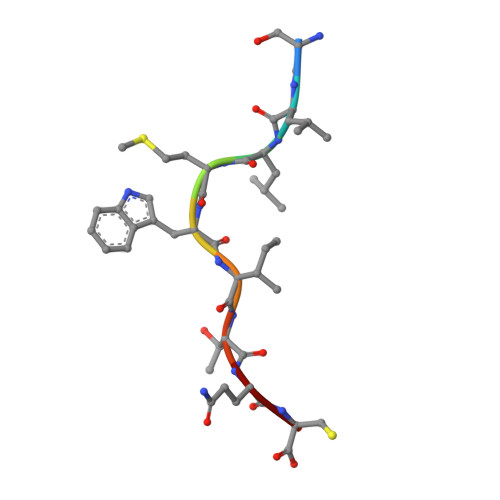
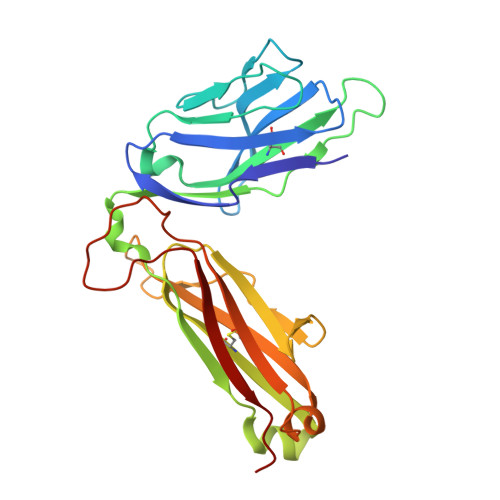
Crystal structures of high affinity human T-cell receptors bound to peptide major histocompatibility complex reveal native diagonal binding geometry
Sami, M., Rizkallah, P.J., Dunn, S., Molloy, P., Moysey, R., Vuidepot, A., Baston, E., Todorov, P., Yi, L., Gao, F., Boulter, J.M., Jakobsen, B.K.(2007) Protein Eng Des Sel 20: 397-403
- PubMed: 17644531
- DOI: https://doi.org/10.1093/protein/gzm033
- Primary Citation of Related Structures:
2P5E, 2P5W, 2PYE, 2PYF - PubMed Abstract:
Naturally selected T-cell receptors (TCRs) are characterised by low binding affinities, typically in the range 1-100 microM. Crystal structures of syngeneic TCRs bound to peptide major histocompatibility complex (pMHC) antigens exhibit a conserved mode of binding characterised by a distinct diagonal binding geometry, with poor shape complementarity (SC) between receptor and ligand. Here, we report the structures of three in vitro affinity enhanced TCRs that recognise the pMHC tumour epitope NY-ESO(157-165) (SLLMWITQC). These crystal structures reveal that the docking mode for the high affinity TCRs is identical to that reported for the parental wild-type TCR, with only subtle changes in the mutated complementarity determining regions (CDRs) that form contacts with pMHC; both CDR2 and CDR3 mutations act synergistically to improve the overall affinity. Comparison of free and bound TCR structures for both wild-type and a CDR3 mutant reveal an induced fit mechanism arising from restructuring of CDR3 loops which allows better peptide binding. Overall, an increased interface area, improved SC and additional H-bonding interactions are observed, accounting for the increase in affinity. Most notably, there is a marked increase in the SC for the central methionine and tryptophan peptide motif over the native TCR.
Organizational Affiliation:
Avidex Limited (subsidiary of Medigene Ag), 57c Milton Park, Abingdon, Oxon OX14 4RX, UK.