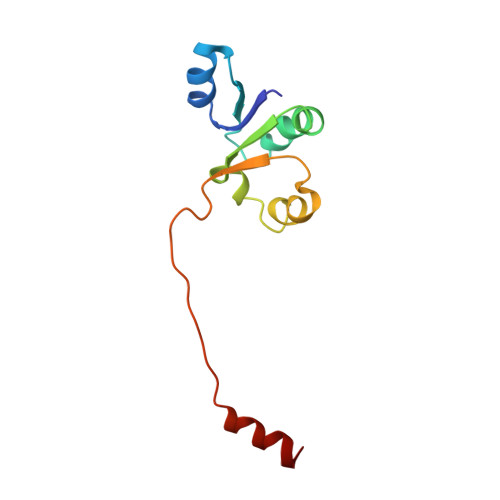
NMR solution structure of subunit F of the methanogenic A1AO adenosine triphosphate synthase and its interaction with the nucleotide-binding subunit B.
Gayen, S., Vivekanandan, S., Biukovic, G., Gruber, G., Yoon, H.S.(2007) Biochemistry 46: 11684-11694
- PubMed: 17910473
- DOI: https://doi.org/10.1021/bi701102n
- Primary Citation of Related Structures:
2OV6 - PubMed Abstract:
The A1AO adenosine triphosphate (ATP) synthase from archaea uses the ion gradients generated across the membrane sector (AO) to synthesize ATP in the A3B3 domain of the A1 sector. The energy coupling between the two active domains occurs via the so-called stalk part(s), to which the 12 kDa subunit F does belong. Here, we present the solution structure of the F subunit of the A1AO ATP synthase from Methanosarcina mazei Gö1. Subunit F exhibits a distinct two-domain structure, with the N-terminal having 78 residues and residues 79-101 forming the flexible C-terminal part. The well-ordered N-terminal domain is composed of a four-stranded parallel beta-sheet structure and three alpha-helices placed alternately. The two domains are loosely associated with more flexibility relative to each other. The flexibility of the C-terminal domain is further confirmed by dynamics studies. In addition, the affinity of binding of mutant subunit F, with a substitution of Trp100 against Tyr and Ile at the very C-terminal end, to the nucleotide-binding subunit B was determined quantitatively using the fluorescence signals of natural subunit B (Trp430). Finally, the arrangement of subunit F within the complex is presented.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551.