Crystal structure of CD59: implications for molecular recognition of the complement proteins C8 and C9 in the membrane-attack complex.
Huang, Y., Fedarovich, A., Tomlinson, S., Davies, C.(2007) Acta Crystallogr D Biol Crystallogr 63: 714-721
- PubMed: 17505110
- DOI: https://doi.org/10.1107/S0907444907015557
- Primary Citation of Related Structures:
2OFS - PubMed Abstract:
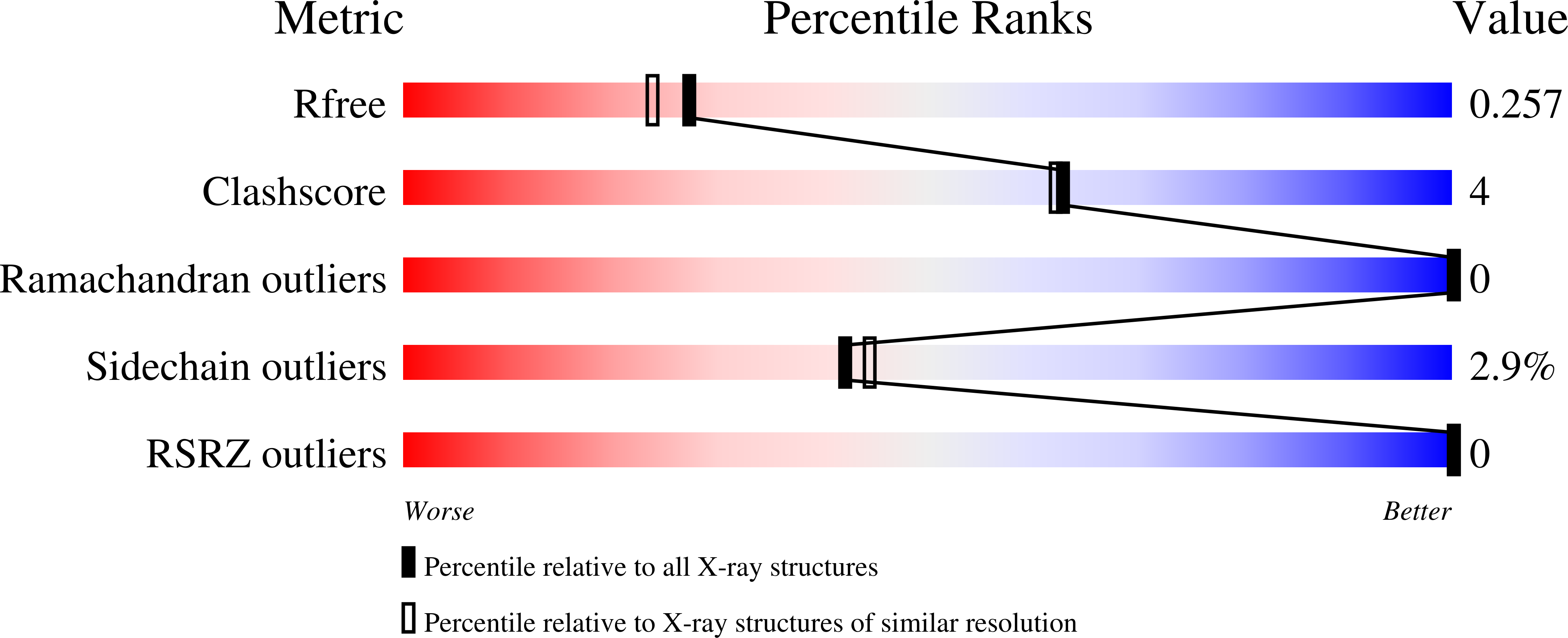
Human CD59 is a small membrane-bound glycoprotein that functions as an inhibitor of the membrane-attack complex (MAC) of the complement system by binding the complement proteins C8 and C9. The crystal structure of a soluble construct of CD59 has been determined to 2.1 A resolution. When compared with previous models of CD59 determined using NMR, some interesting differences are noted, including the position of helix alpha1, which contributes to the binding surface for C8 and C9. Interestingly, the crystal structure superimposes more closely with an updated NMR model of CD59 that was produced using Monte Carlo minimization, including helix alpha1. Mapping of mutations associated with enhanced or lowered inhibitory function of CD59 show the binding region to be located in a crevice between alpha1 and a three-stranded beta-sheet, as has been identified previously. Residues in the core of this region are well ordered in the electron density, in part owing to a network of stabilizing covalent and noncovalent interactions, and manifest an interesting 'striped' distribution of hydrophobic and basic residues. Docking of the same peptide that was modeled previously into the NMR structure shows that Arg55, which has been postulated to exist in 'open' and 'closed' positions, is intermediate in position between these two and is well placed to contact the peptide. Further clues regarding how CD59 interacts with small peptides arise from the crystal packing of this structure, which shows that a symmetry-related loop comprising residues 20-24 occupies a spatially similar position to the modeled peptide. This higher resolution structure of CD59 will facilitate a more precise dissection of its interactions with C8 and C9 and thus increase the likelihood of designing enhanced CD59-based therapeutics.
Organizational Affiliation:
Department of Microbiology and Immunology, Medical University of South Carolina, Charleston, SC 29425, USA.