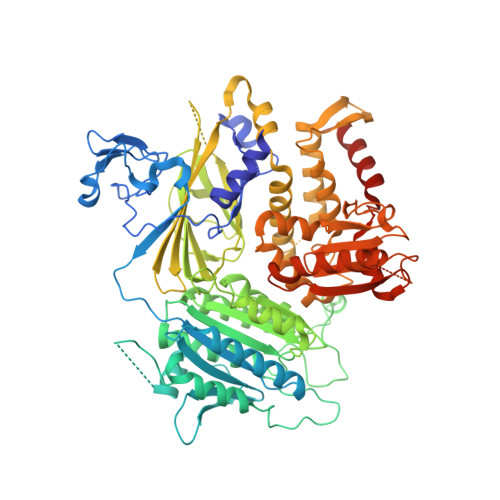
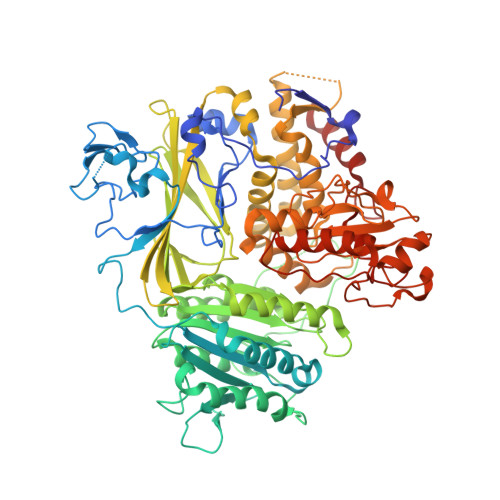
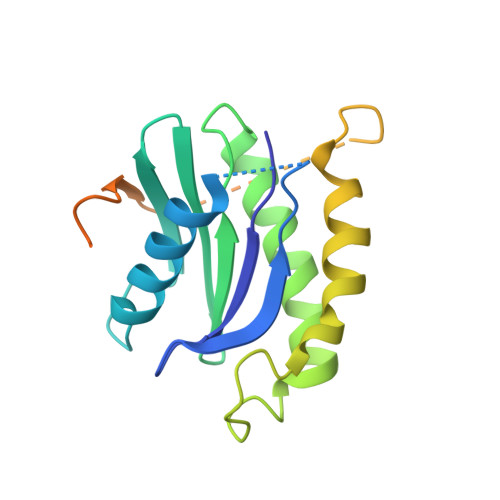
The Transport Signal on Sec22 for Packaging into COPII-Coated Vesicles is a Conformational Epitope
Mancias, J.D., Goldberg, J.(2007) Mol Cell 26: 403-414
- PubMed: 17499046
- DOI: https://doi.org/10.1016/j.molcel.2007.03.017
- Primary Citation of Related Structures:
2NUP, 2NUT - PubMed Abstract:
The mechanism of cargo concentration into ER-derived vesicles involves interactions between the COPII vesicular coat complex and cargo transport signals--peptide sequences of 10-15 residues. The SNARE protein Sec22 contains a signal that binds the COPII subcomplex Sec23/24 and specifies its endoplasmic reticulum (ER) exit as an unassembled SNARE. The 200 kDa crystal structure of Sec22 bound to Sec23/24 reveals that the transport signal is a folded epitope rather than a conventional short peptide sequence. The NIE segment of the SNARE motif folds against the N-terminal longin domain, and this closed form of Sec22 binds at the Sec23/24 interface. Thus, COPII recognizes unassembled Sec22 via a folded epitope, whereas Sec22 assembly into SNARE complexes would mask the NIE segment. The concept of a conformational epitope as a transport signal suggests packaging mechanisms in which a coat is sensitive to the folded state of a cargo protein or the assembled state of a multiprotein complex.
Organizational Affiliation:
Howard Hughes Medical Institute, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, USA.