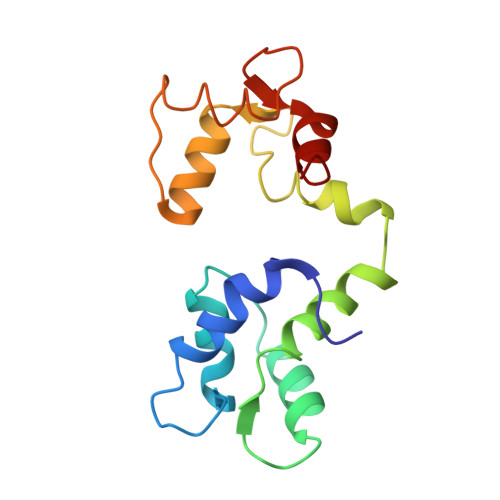
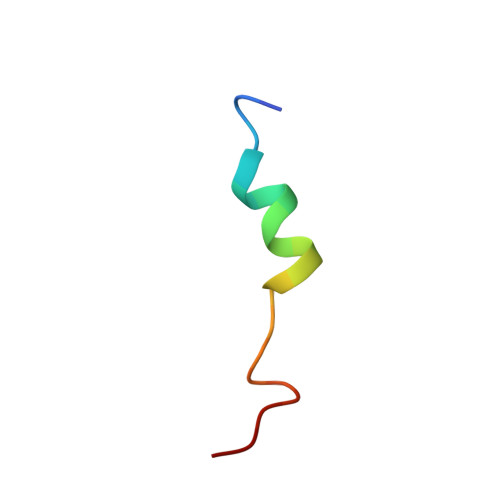
Structural Studies of a Complex Between Endothelial Nitric Oxide Synthase and Calmodulin at Physiological Calcium Concentration.
Piazza, M., Dieckmann, T., Guillemette, J.G.(2016) Biochemistry 55: 5962-5971
- PubMed: 27696828
- DOI: https://doi.org/10.1021/acs.biochem.6b00821
- Primary Citation of Related Structures:
2N8J - PubMed Abstract:
The small acidic protein calmodulin (CaM) serves as a Ca 2+ sensor and control element for many enzymes including nitric oxide synthase (NOS) enzymes that play major roles in key physiological and pathological processes. CaM binding causes a conformational change in NOS to allow for the electron transfer between the reductase and oxygenase domains through a process that is thought to be highly dynamic. In this report, NMR spectroscopy was used to determine the solution structure of the endothelial NOS (eNOS) peptide in complex with CaM at the lowest Ca 2+ concentration (225 nM) required for CaM to bind to eNOS and corresponds to a physiological elevated Ca 2+ level found in mammalian cells. Under these conditions, the CaM-eNOS complex has a Ca 2+ -replete C-terminal lobe bound to the eNOS peptide and a Ca 2+ free N-terminal lobe loosely associated with the eNOS peptide. With increasing Ca 2+ concentration, the binding of Ca 2+ by the N-lobe of CaM results in a stronger interaction with the C-terminal region of the eNOS peptide and increased α-helical structure of the peptide that may be part of the mechanism resulting in electron transfer from the FMN to the heme in the oxygenase domain of the enzyme. Surface plasmon resonance studies performed under the same conditions show Ca 2+ concentration-dependent binding kinetics were consistent with the NMR structural results. This investigation shows that structural studies performed under more physiological relevant conditions provide information on subtle changes in structure that may not be apparent when experiments are performed in excess Ca 2+ concentrations.
Organizational Affiliation:
Department of Chemistry, University of Waterloo , Waterloo, Ontario N2L 3G1, Canada.