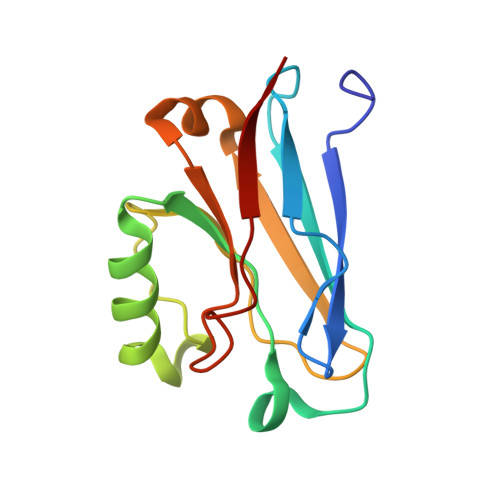
The solution structure of the soluble form of the lipid-modified azurin from Neisseria gonorrhoeae, the electron donor of cytochrome c peroxidase.
Nobrega, C.S., Saraiva, I.H., Carreira, C., Devreese, B., Matzapetakis, M., Pauleta, S.R.(2016) Biochim Biophys Acta 1857: 169-176
- PubMed: 26589091
- DOI: https://doi.org/10.1016/j.bbabio.2015.11.006
- Primary Citation of Related Structures:
2N0M - PubMed Abstract:
Neisseria gonorrhoeae colonizes the genitourinary track, and in these environments, especially in the female host, the bacteria are subjected to low levels of oxygen, and reactive oxygen and nitrosyl species. Here, the biochemical characterization of N. gonorrhoeae Laz is presented, as well as, the solution structure of its soluble domain determined by NMR. N. gonorrhoeae Laz is a type 1 copper protein of the azurin-family based on its spectroscopic properties and structure, with a redox potential of 277±5 mV, at pH7.0, that behaves as a monomer in solution. The globular Laz soluble domain adopts the Greek-key motif, with the copper center located at one end of the β-barrel coordinated by Gly48, His49, Cys113, His118 and Met122, in a distorted trigonal geometry. The edge of the His118 imidazole ring is water exposed, in a surface that is proposed to be involved in the interaction with its redox partners. The heterologously expressed Laz was shown to be a competent electron donor to N. gonorrhoeae cytochrome c peroxidase. This is an evidence for its involvement in the mechanism of protection against hydrogen peroxide generated by neighboring lactobacilli in the host environment.
Organizational Affiliation:
UCIBIO, REQUIMTE, Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, Campus da Caparica, 2829-516 Caparica, Portugal.