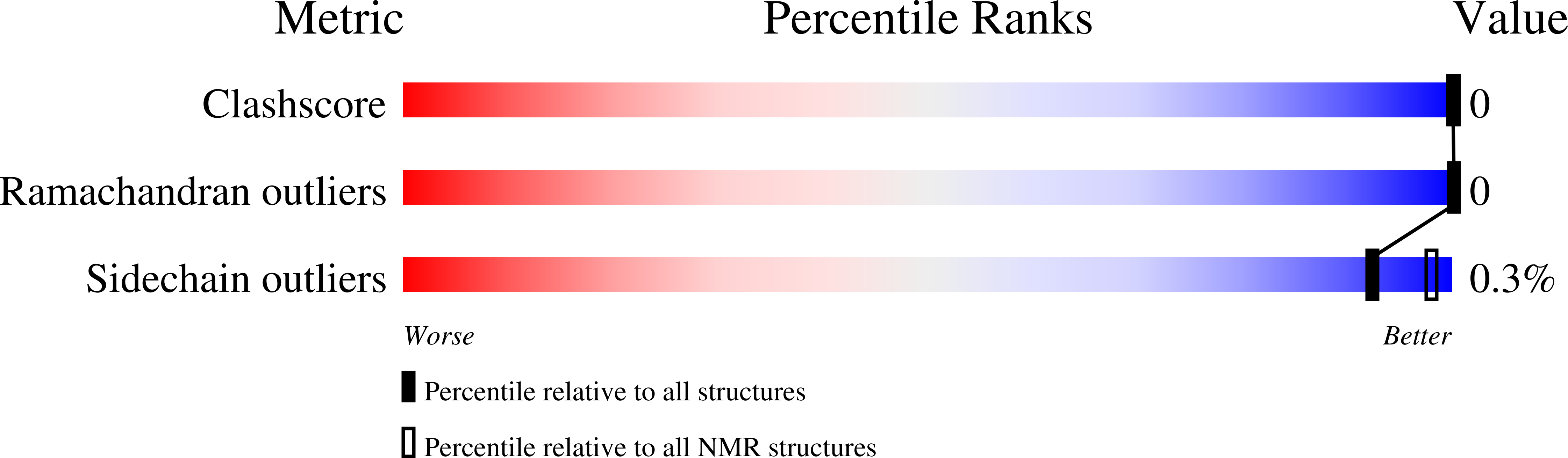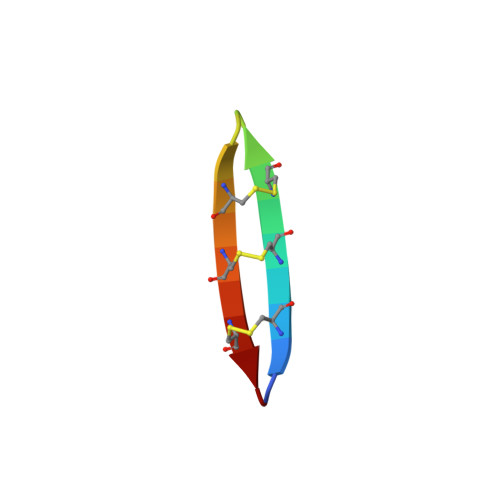Structural characterization of the cyclic cystine ladder motif of theta-defensins.
Conibear, A.C., Rosengren, K.J., Harvey, P.J., Craik, D.J.(2012) Biochemistry 51: 9718-9726
- PubMed: 23148585
- DOI: https://doi.org/10.1021/bi301363a
- Primary Citation of Related Structures:
2LYE, 2LYF, 2LZI - PubMed Abstract:
The θ-defensins are, to date, the only known ribosomally synthesized cyclic peptides in mammals, and they have promising antimicrobial bioactivities. The characteristic structural motif of the θ-defensins is the cyclic cystine ladder, comprising a cyclic peptide backbone and three parallel disulfide bonds. In contrast to the cyclic cystine knot, which characterizes the plant cyclotides, the cyclic cystine ladder has not been as well described as a structural motif. Here we report the solution structures and nuclear magnetic resonance relaxation properties in aqueous solution of three representative θ-defensins from different species. Our data suggest that the θ-defensins are more rigid and structurally defined than previously thought. In addition, all three θ-defensins were found to self-associate in aqueous solution in a concentration-dependent and reversible manner, a property that might have a role in their mechanism of action. The structural definition of the θ-defensins and the cyclic cystine ladder will help to guide exploitation of these molecules as structural frameworks for the design of peptide drugs.
Organizational Affiliation:
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD 4072, Australia.