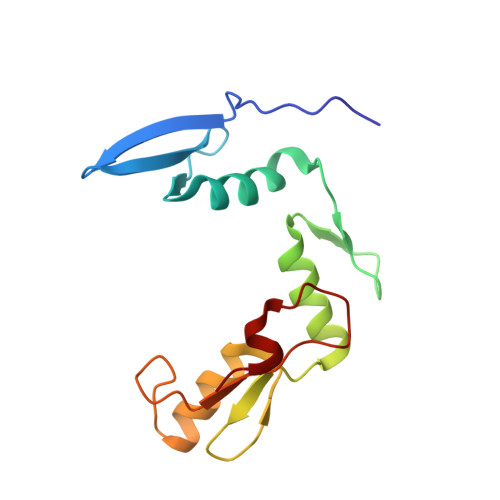
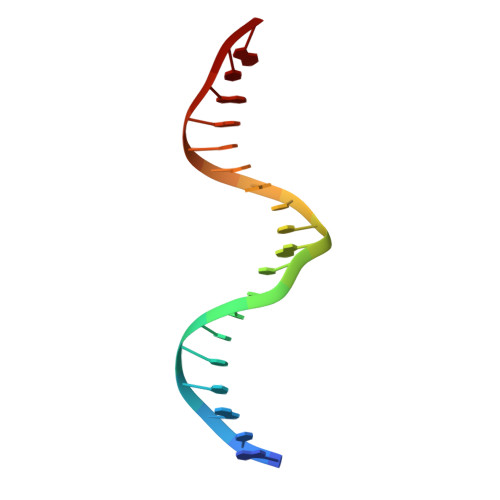
Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Buck-Koehntop, B.A., Stanfield, R.L., Ekiert, D.C., Martinez-Yamout, M.A., Dyson, H.J., Wilson, I.A., Wright, P.E.(2012) Proc Natl Acad Sci U S A 109: 15229-15234
- PubMed: 22949637
- DOI: https://doi.org/10.1073/pnas.1213726109
- Primary Citation of Related Structures:
2LT7, 4F6M, 4F6N - PubMed Abstract:
Methylation of CpG dinucleotides in DNA is a common epigenetic modification in eukaryotes that plays a central role in maintenance of genome stability, gene silencing, genomic imprinting, development, and disease. Kaiso, a bifunctional Cys(2)His(2) zinc finger protein implicated in tumor-cell proliferation, binds to both methylated CpG (mCpG) sites and a specific nonmethylated DNA motif (TCCTGCNA) and represses transcription by recruiting chromatin remodeling corepression machinery to target genes. Here we report structures of the Kaiso zinc finger DNA-binding domain in complex with its nonmethylated, sequence-specific DNA target (KBS) and with a symmetrically methylated DNA sequence derived from the promoter region of E-cadherin. Recognition of specific bases in the major groove of the core KBS and mCpG sites is accomplished through both classical and methyl CH···O hydrogen-bonding interactions with residues in the first two zinc fingers, whereas residues in the C-terminal extension following the third zinc finger bind in the opposing minor groove and are required for high-affinity binding. The C-terminal region is disordered in the free protein and adopts an ordered structure upon binding to DNA. The structures of these Kaiso complexes provide insights into the mechanism by which a zinc finger protein can recognize mCpG sites as well as a specific, nonmethylated regulatory DNA sequence.
Organizational Affiliation:
Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.