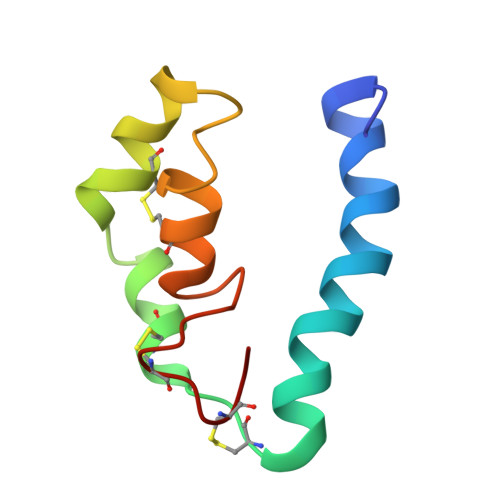
NMR structure of a fungal virulence factor reveals structural homology with mammalian saposin B.
Beck, M.R., Dekoster, G.T., Cistola, D.P., Goldman, W.E.(2009) Mol Microbiol 72: 344-353
- PubMed: 19298372
- DOI: https://doi.org/10.1111/j.1365-2958.2009.06647.x
- Primary Citation of Related Structures:
2JV7 - PubMed Abstract:
The fungal protein CBP (calcium binding protein) is a known virulence factor with an unknown virulence mechanism. The protein was identified based on its ability to bind calcium and its prevalence as Histoplasma capsulatum's most abundant secreted protein. However, CBP has no sequence homology with other CBPs and contains no known calcium binding motifs. Here, the NMR structure of CBP reveals a highly intertwined homodimer and represents the first atomic level NMR model of any fungal virulence factor. Each CBP monomer is comprised of four alpha-helices that adopt the saposin fold, characteristic of a protein family that binds to membranes and lipids. This structural homology suggests that CBP functions as a lipid binding protein, potentially interacting with host glycolipids in the phagolysosome of host cells.
Organizational Affiliation:
Department of Molecular Microbiology, Washington University in St Louis, St Louis, MO, USA.