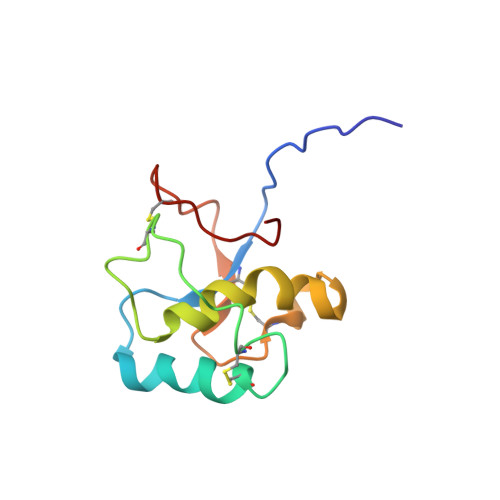
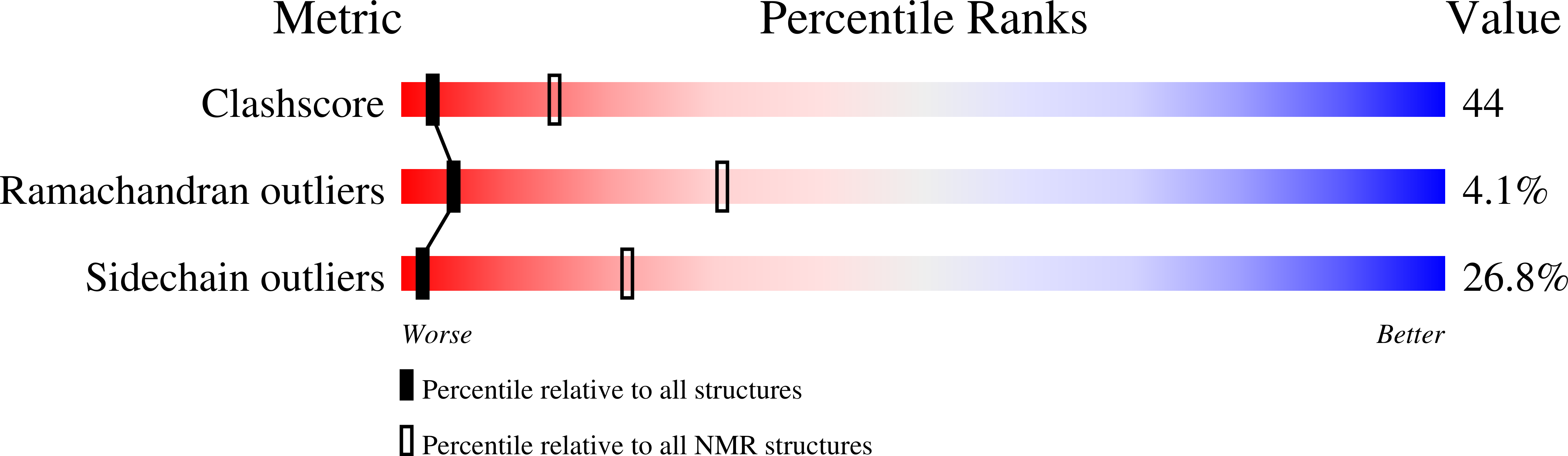Solution structure of the C-terminal domain of Ole e 9, a major allergen of olive pollen
Trevino, M.A., Palomares, O., Castrillo, I., Villalba, M., Rodriguez, R., Rico, M., Santoro, J., Bruix, M.(2008) Protein Sci 17: 371-376
- PubMed: 18096638
- DOI: https://doi.org/10.1110/ps.073230008
- Primary Citation of Related Structures:
2JON - PubMed Abstract:
Ole e 9 is an olive pollen allergen belonging to group 2 of pathogenesis-related proteins. The protein is composed of two immunological independent domains: an N-terminal domain (NtD) with 1,3-beta-glucanase activity, and a C-terminal domain (CtD) that binds 1,3-beta-glucans. We have determined the three-dimensional structure of CtD-Ole e 9 (101 amino acids), which consists of two parallel alpha-helices forming an angle of approximately 55 degrees , a small antiparallel beta-sheet with two short strands, and a 3-10 helix turn, all connected by long coil segments, resembling a novel type of folding among allergens. Two regions surrounded by aromatic residues (F49, Y60, F96, Y91 and Y31, H68, Y65, F78) have been localized on the protein surface, and a role for sugar binding is suggested. The epitope mapping of CtD-Ole e 9 shows that B-cell epitopes are mainly located on loops, although some of them are contained in secondary structural elements. Interestingly, the IgG and IgE epitopes are contiguous or overlapped, rather than coincident. The three-dimensional structure of CtD-Ole e 9 might help to understand the underlying mechanism of its biochemical function and to determine possible structure-allergenicity relationships.
Organizational Affiliation:
Departamento de Espectroscopía y Estructura Molecular, Instituto de Química Física Rocasolano, CSIC, 28006 Madrid, Spain.