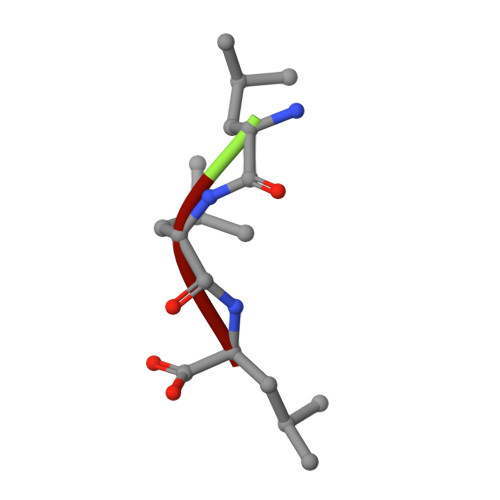
Experimental evidence for a metallohydrolase mechanism in which the nucleophile is not delivered by a metal ion: EPR spectrokinetic and structural studies of aminopeptidase from Vibrio proteolyticus
Kumar, A., Periyannan, G.R., Narayanan, B., Kittell, A.W., Kim, J.-J., Bennett, B.(2007) Biochem J 403: 527-536
- PubMed: 17238863
- DOI: https://doi.org/10.1042/BJ20061591
- Primary Citation of Related Structures:
2IQ6 - PubMed Abstract:
Metallohydrolases catalyse some of the most important reactions in biology and are targets for numerous chemotherapeutic agents designed to combat bacterial infectivity, antibiotic resistance, HIV infectivity, tumour growth, angiogenesis and immune disorders. Rational design of inhibitors of these enzymes with chemotherapeutic potential relies on detailed knowledge of the catalytic mechanism. The roles of the catalytic transition ions in these enzymes have long been assumed to include the activation and delivery of a nucleophilic hydroxy moiety. In the present study, catalytic intermediates in the hydrolysis of L-leucyl-L-leucyl-L-leucine by Vibrio proteolyticus aminopeptidase were characterized in spectrokinetic and structural studies. Rapid-freeze-quench EPR studies of reaction products of L-leucyl-L-leucyl-L-leucine and Co(II)-substituted aminopeptidase, and comparison of the EPR data with those from structurally characterized complexes of aminopeptidase with inhibitors, indicated the formation of a catalytically competent post-Michaelis pre-transition state intermediate with a structure analogous to that of the inhibited complex with bestatin. The X-ray crystal structure of an aminopeptidase-L-leucyl-L-leucyl-L-leucine complex was also analogous to that of the bestatin complex. In these structures, no water/hydroxy group was observed bound to the essential metal ion. However, a water/hydroxy group was clearly identified that was bound to the metal-ligating oxygen atom of Glu152. This water/hydroxy group is proposed as a candidate for the active nucleophile in a novel metallohydrolase mechanism that shares features of the catalytic mechanisms of aspartic proteases and of B2 metallo-beta-lactamases. Preliminary studies on site-directed variants are consistent with the proposal. Other features of the structure suggest roles for the dinuclear centre in geometrically and electrophilically activating the substrate.
Organizational Affiliation:
Department of Biophysics, Medical College of Wisconsin, Milwaukee, WI 53226-0509, USA.