The Central Cavity from the (Alpha/Alpha)(6) Barrel Structure of Anabaena sp. CH1 N-Acetyl-d-glucosamine 2-Epimerase Contains Two Key Histidine Residues for Reversible Conversion.
Lee, Y.C., Wu, H.M., Chang, Y.N., Wang, W.C., Hsu, W.H.(2007) J Mol Biol 367: 895-908
- PubMed: 17292397
- DOI: https://doi.org/10.1016/j.jmb.2006.11.001
- Primary Citation of Related Structures:
2GZ6 - PubMed Abstract:
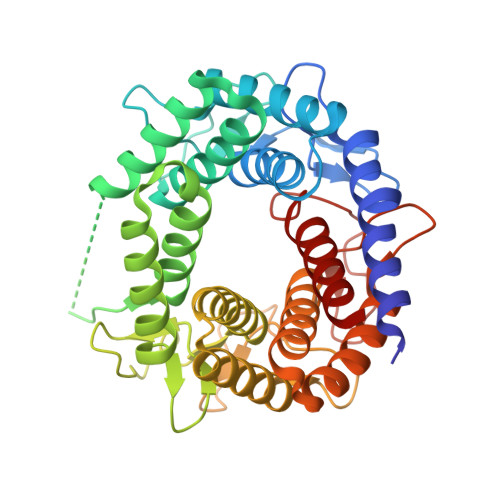
N-acetyl-D-glucosamine 2-epimerase (GlcNAc 2-epimerase) catalyzes the reversible epimerization between N-acetyl-D-glucosamine (GlcNAc) and N-acetyl-D-mannosamine (ManNAc). We report here the 2.0 A resolution crystal structure of the GlcNAc 2-epimerase from Anabaena sp. CH1. The structure demonstrates an (alpha/alpha)(6) barrel fold, which shows structural homology with porcine GlcNAc 2-epimerase as well as a number of glycoside hydrolase enzymes and other sugar-metabolizing enzymes. One side of the barrel structure consists of short loops involved in dimer interactions. The other side of the barrel structure is comprised of long loops containing six short beta-sheets, which enclose a putative central active-site pocket. Site-directed mutagenesis of conserved residues near the N-terminal region of the inner alpha helices shows that R57, H239, E308, and H372 are strictly required for activity. E242 and R375 are also essential in catalysis. Based on the structure and kinetic analysis, H239 and H372 may serve as the key active site acid/base catalysts. These results suggest that the (alpha/alpha)(6) barrel represents a steady fold for presenting active-site residues in a cleft at the N-terminal ends of the inner alpha helices, thus forming a fine-tuned catalytic site in GlcNAc 2-epimerase.
Organizational Affiliation:
Institute of Molecular Biology, National Chung Hsing University, Taichung, Taiwan.