Non-Canonical Yxxg-Phi Endocytic Motifs: Recognition by Ap2 and Preferential Utilization in P2X4 Receptors.
Royle, S.J., Qureshi, O.S., Bobanovic, L.K., Evans, P.R., Owen, D.J., Murrell-Lagnado, R.D.(2005) J Cell Sci 118: 3073
- PubMed: 15985462
- DOI: https://doi.org/10.1242/jcs.02451
- Primary Citation of Related Structures:
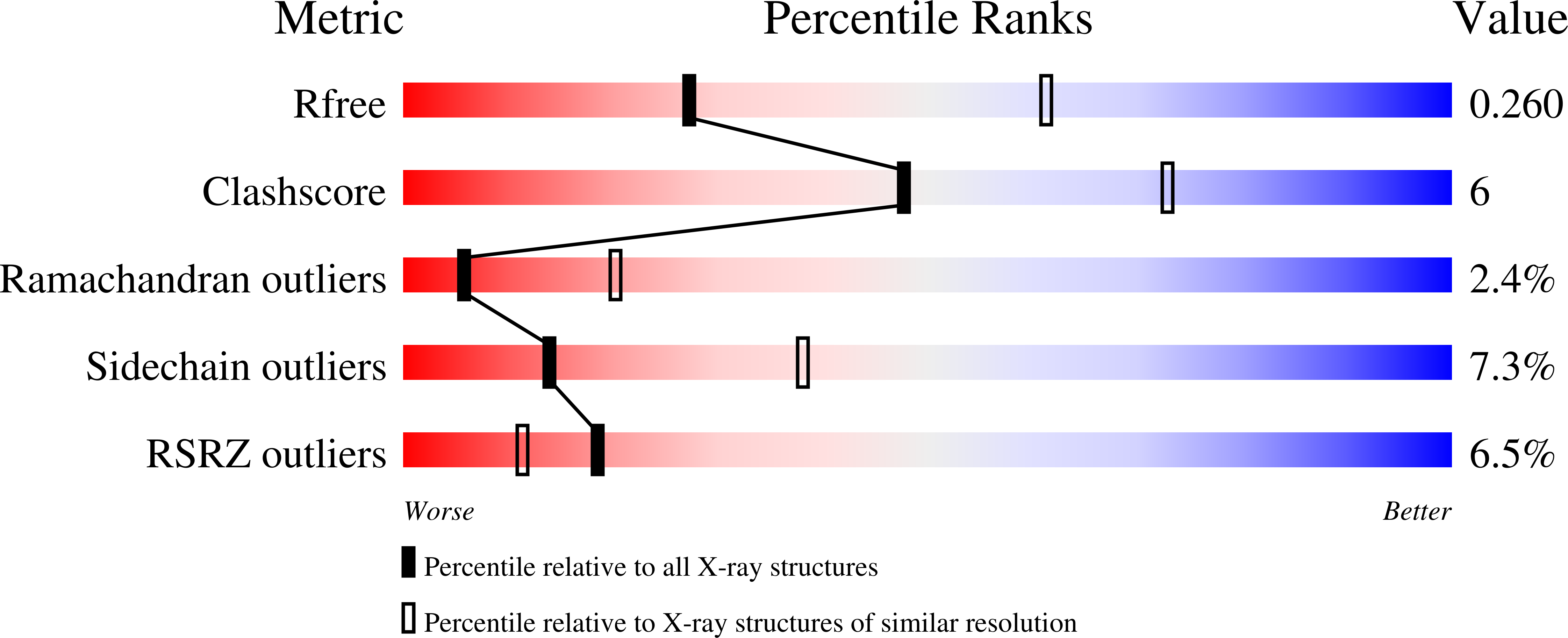
2BP5 - PubMed Abstract:
During clathrin-mediated endocytosis, proteins on the cell surface are selected for inclusion in clathrin-coated vesicles by clathrin adaptors, mainly the adaptor complex AP2. The P2X4 subtype of ATP-gated ion channel has in its C-terminus two putative endocytic motifs: a canonical YXXPhi motif and a non-canonical YXXGPhi motif (YEQGL). We demonstrate that endocytosis of P2X4 receptors is mediated preferentially by the YXXGPhi motif because the YXXPhi motif is inaccessible to AP2 owing to the structure of the channel. The crystal structure of a complex between residues 160-435 of the mu2 subunit of AP2 and a P2X4 C-terminal peptide showed that the YEQGL motif binds to mu2 at the same site as YXXPhi motifs. Y and Phi residues are accommodated in the same hydrophobic pockets in mu2 with the extra residue between them being accommodated by changes in the peptide's backbone configuration, when compared to YXXPhi motifs. These data demonstrate that the family of potential tyrosine-based endocytic signals must be expanded to include motifs with an additional glycine at Y+3 (YXXGPhi).
Organizational Affiliation:
Department of Pharmacology, University of Cambridge, Tennis Court Road, Cambridge CB2 1PD, UK.