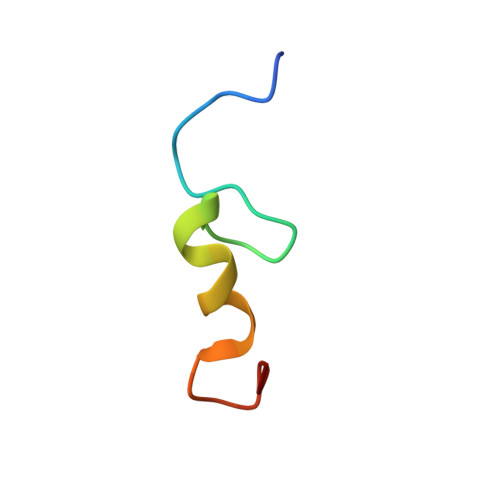
NMR structure of the mengovirus Leader protein zinc-finger domain.
Cornilescu, C.C., Porter, F.W., Zhao, K.Q., Palmenberg, A.C., Markley, J.L.(2008) FEBS Lett 582: 896-900
- PubMed: 18291103
- DOI: https://doi.org/10.1016/j.febslet.2008.02.023
- Primary Citation of Related Structures:
2BAI - PubMed Abstract:
The Leader protein is a defining feature of picornaviruses from the Cardiovirus genus. This protein was recently shown to inhibit cellular nucleocytoplasmic transport through an activity mapped to its zinc-binding region. Here we report the three-dimensional solution structure determined by nuclear magnetic resonance (NMR) spectroscopy of this domain (residues 5-28) from mengovirus. The domain forms a CHCC zinc-finger with a fold comprising a beta-hairpin followed by a short alpha-helix that can adopt two different conformations. This structure is divergent from those of other eukaryotic zinc-fingers and instead resembles motifs found in a group of DNA-binding proteins from Archaea.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin-Madison, Madison, WI 53706-1544, USA. cclaudia@nmrfam.wisc.edu