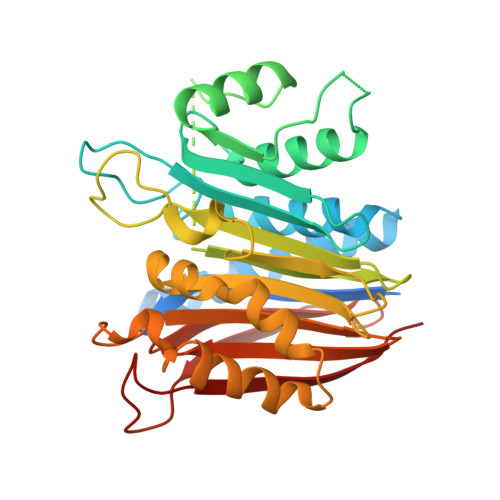
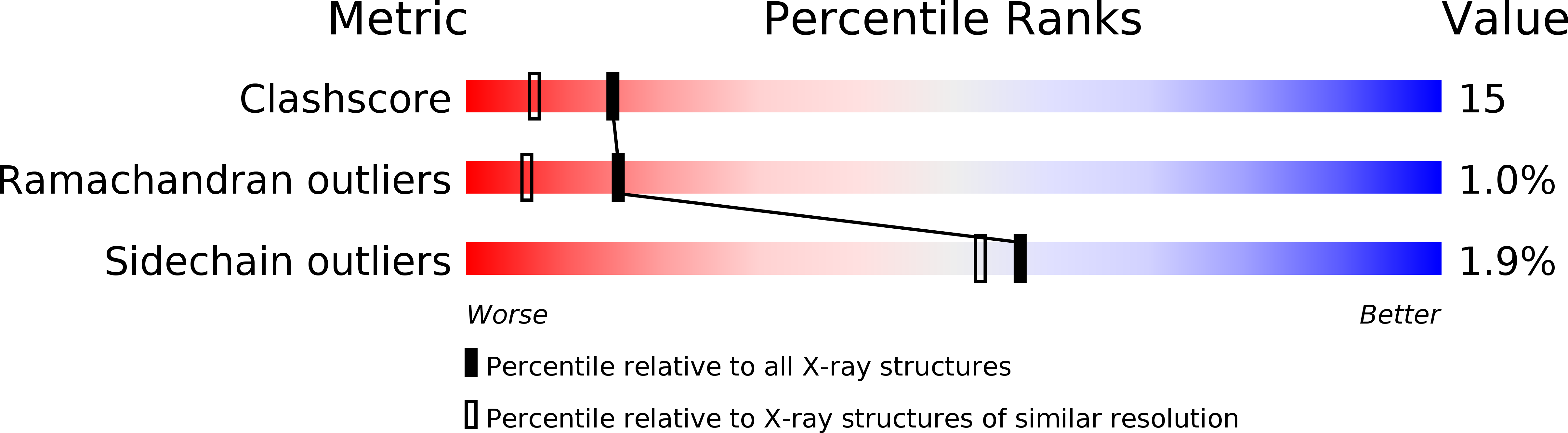Crystal Structure of Human Taspase1, a Crucial Protease Regulating the Function of MLL.
Khan, J.A., Dunn, B.M., Tong, L.(2005) Structure 13: 1443-1452
- PubMed: 16216576
- DOI: https://doi.org/10.1016/j.str.2005.07.006
- Primary Citation of Related Structures:
2A8I, 2A8J, 2A8L, 2A8M - PubMed Abstract:
Taspase1 catalyzes the proteolytic processing of the mixed lineage leukemia (MLL) nuclear protein, which is required for maintaining Hox gene expression patterns. Chromosomal translocations of the MLL gene are associated with leukemia in infants. Taspase1, a threonine aspartase, is a member of the type 2 asparaginase family, but is the only protease in this family. We report here the crystal structures of human activated Taspase1 and its proenzyme, as well as the characterization of the effects of mutations in the active site region using a newly developed fluorogenic assay. The structure of Taspase1 has significant differences from other asparaginases, especially near the active site. Mutation of the catalytic nucleophile, Thr234, abolishes autocatalytic processing in cis but does not completely block proteolysis in trans. The structure unexpectedly showed the binding of a chloride ion in the active site, and our kinetic studies confirm that chlorides ions are inhibitors of this enzyme at physiologically relevant concentrations.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York 10027, USA.