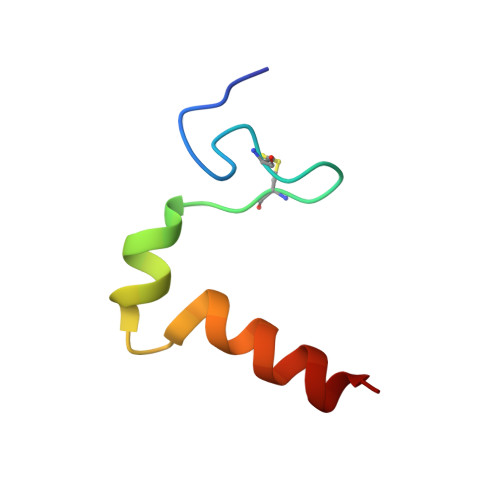
Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide curvacin A
Haugen, H.S., Fimland, G., Nissen-Meyer, J., Kristiansen, P.E.(2005) Biochemistry 44: 16149-16157
- PubMed: 16331975
- DOI: https://doi.org/10.1021/bi051215u
- Primary Citation of Related Structures:
2A2B - PubMed Abstract:
The 3D structure of the membrane-permeabilizing 41-mer pediocin-like antimicrobial peptide curvacin A produced by lactic acid bacteria has been studied by NMR spectroscopy. In DPC micelles, the cationic and hydrophilic N-terminal half of the peptide forms an S-shaped beta-sheet-like domain stabilized by a disulfide bridge and a few hydrogen bonds. This domain is followed by two alpha-helices: a hydrophilic 6-mer helix between residues 19 and 24 and an amphiphilic/hydrophobic 11-mer helix between residues 29 and 39. There are two hinges in the peptide, one at residues 16-18 between the N-terminal S-shaped beta-sheet-like structure and the central 6-mer helix and one at residues 26-28 between the central helix and the 11-mer C-terminal helix. The latter helix is the only amphiphilic/hydrophobic part of the peptide and is thus presumably the part that penetrates into the hydrophobic phase of target-cell membranes. The hinge between the two helices may introduce the flexibility that allows the helix to dip into membranes. The helix-hinge-helix structure in the C-terminal half of curvacin A clearly distinguishes this peptide from the other pediocin-like peptides whose structures have been analyzed and suggests that curvacin A along with the structural homologues enterocin P and carnobacteriocin BM1 belong to a subgroup of the pediocin-like family of antimicrobial peptides.
Organizational Affiliation:
Department of Molecular Biosciences, University of Oslo, Pb. 1041 Blindern, 0316 Oslo, Norway.