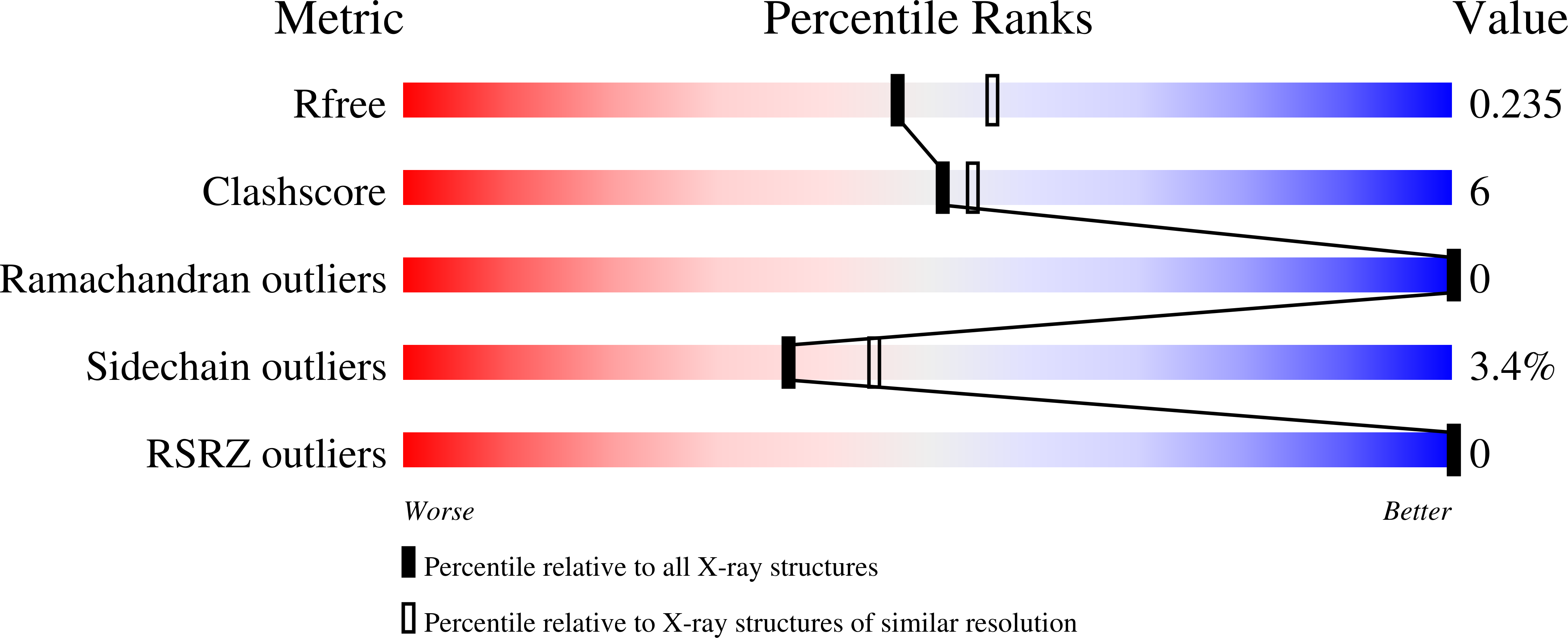
Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
Orru, R., Dudek, H.M., Martinoli, C., Torres Pazmino, D.E., Royant, A., Weik, M., Fraaije, M.W., Mattevi, A.(2011) J Biol Chem 286: 29284
- PubMed: 21697090
- DOI: https://doi.org/10.1074/jbc.M111.255075
- Primary Citation of Related Structures:
2YLR, 2YLS, 2YLT, 2YLW, 2YLX, 2YLZ, 2YM1, 2YM2 - PubMed Abstract:
Baeyer-Villiger monooxygenases catalyze the oxidation of carbonylic substrates to ester or lactone products using NADPH as electron donor and molecular oxygen as oxidative reactant. Using protein engineering, kinetics, microspectrophotometry, crystallography, and intermediate analogs, we have captured several snapshots along the catalytic cycle which highlight key features in enzyme catalysis. After acting as electron donor, the enzyme-bound NADP(H) forms an H-bond with the flavin cofactor. This interaction is critical for stabilizing the oxygen-activating flavin-peroxide intermediate that results from the reaction of the reduced cofactor with oxygen. An essential active-site arginine acts as anchoring element for proper binding of the ketone substrate. Its positively charged guanidinium group can enhance the propensity of the substrate to undergo a nucleophilic attack by the flavin-peroxide intermediate. Furthermore, the arginine side chain, together with the NADP(+) ribose group, forms the niche that hosts the negatively charged Criegee intermediate that is generated upon reaction of the substrate with the flavin-peroxide. The fascinating ability of Baeyer-Villiger monooxygenases to catalyze a complex multistep catalytic reaction originates from concerted action of this Arg-NADP(H) pair and the flavin subsequently to promote flavin reduction, oxygen activation, tetrahedral intermediate formation, and product synthesis and release. The emerging picture is that these enzymes are mainly oxygen-activating and "Criegee-stabilizing" catalysts that act on any chemically suitable substrate that can diffuse into the active site, emphasizing their potential value as toolboxes for biocatalytic applications.
Organizational Affiliation:
Department of Genetics and Microbiology, University of Pavia, Via Ferrata 1, 27100 Pavia, Italy.