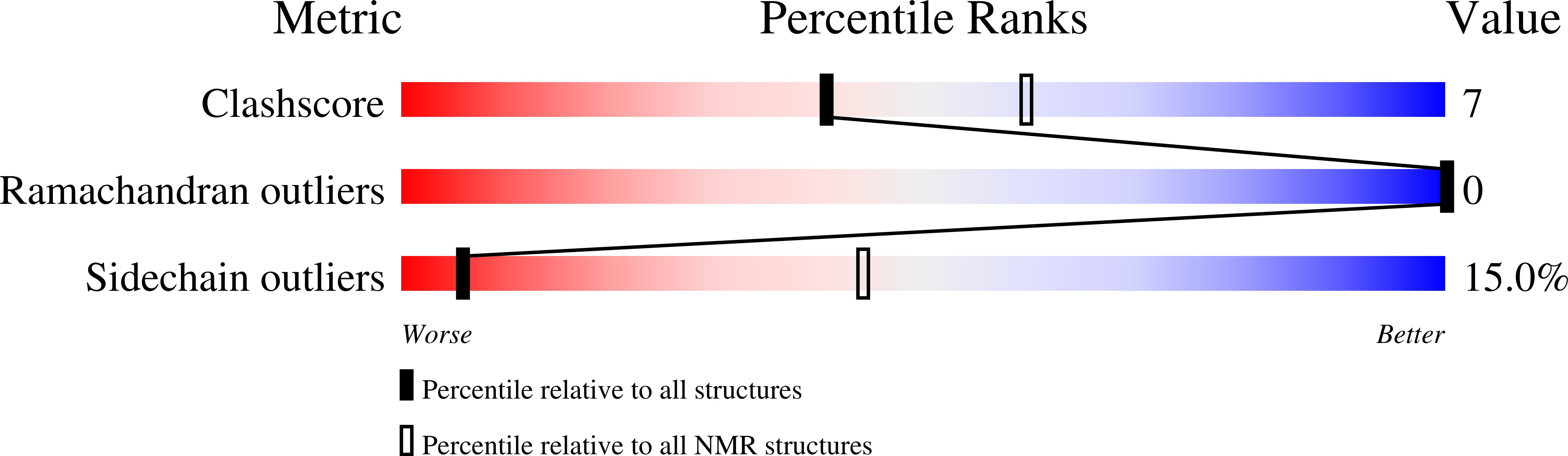Structure and dynamics of helix-0 of the N-BAR domain in lipid micelles and bilayers
Low, C., Weininger, U., Lee, H., Schweimer, K., Neundorf, I., Beck-Sickinger, A.G., Pastor, R.W., Balbach, J.(2008) Biophys J 95: 4315-4323
- PubMed: 18658220
- DOI: https://doi.org/10.1529/biophysj.108.134155
- Primary Citation of Related Structures:
2RMY, 2RND - PubMed Abstract:
Bin/Amphiphysin/Rvs-homology (BAR) domains generate and sense membrane curvature by binding the negatively charged membrane to their positively charged concave surfaces. N-BAR domains contain an N-terminal extension (helix-0) predicted to form an amphipathic helix upon membrane binding. We determined the NMR structure and nano-to-picosecond dynamics of helix-0 of the human Bin1/Amphiphysin II BAR domain in sodium dodecyl sulfate and dodecylphosphocholine micelles. Molecular dynamics simulations of this 34-amino acid peptide revealed electrostatic and hydrophobic interactions with the detergent molecules that induce helical structure formation from residues 8-10 toward the C-terminus. The orientation in the micelles was experimentally confirmed by backbone amide proton exchange. The simulation and the experiment indicated that the N-terminal region is disordered, and the peptide curves to adopted the micelle shape. Deletion of helix-0 reduced tubulation of liposomes by the BAR domain, whereas the helix-0 peptide itself was fusogenic. These findings support models for membrane curving by BAR domains in which helix-0 increases the binding affinity to the membrane and enhances curvature generation.
Organizational Affiliation:
Institut für Physik, Biophysik, Martin-Luther-Universität Halle-Wittenberg, D-06120 Halle (Saale), Germany.