Insights into the role of oligomeric state on the biological activities of crotoxin: crystal structure of a tetrameric phospholipase A2 formed by two isoforms of crotoxin B from Crotalus durissus terrificus venom.
Marchi-Salvador, D.P., Correa, L.C., Magro, A.J., Oliveira, C.Z., Soares, A.M., Fontes, M.R.(2008) Proteins 72: 883-891
- PubMed: 18275084
- DOI: https://doi.org/10.1002/prot.21980
- Primary Citation of Related Structures:
2QOG - PubMed Abstract:
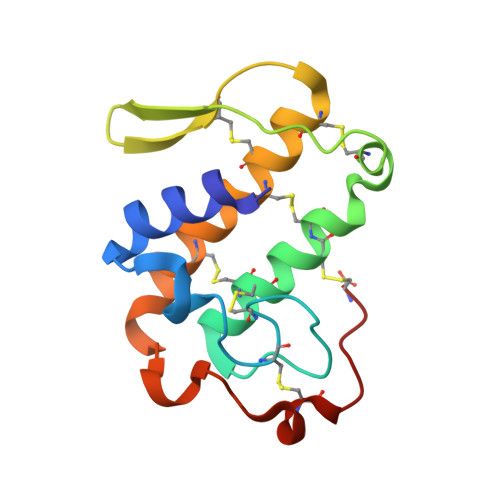
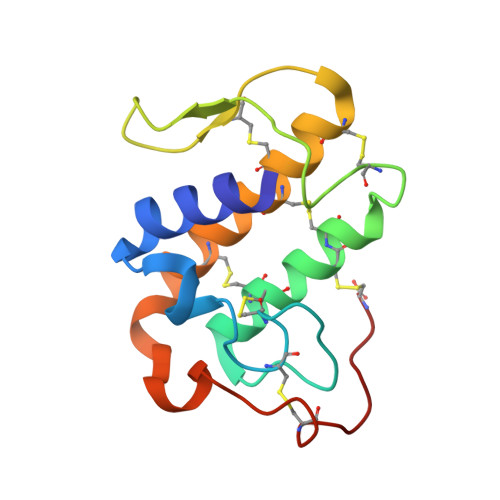
Crotoxin B (CB or Cdt PLA(2)) is a basic Asp49-PLA(2) found in the venom of Crotalus durissus terrificus and it is one of the subunits that constitute the crotoxin (Cro). This heterodimeric toxin, main component of the C. d. terrificus venom, is completed by an acidic, nontoxic, and nonenzymatic component (crotoxin A, CA or crotapotin), and it is related to important envenomation effects such as neurological disorders, myotoxicity, and renal failure. Although Cro has been crystallized since 1938, no crystal structure of this toxin or its subunits is currently available. In this work, the authors present the crystal structure of a novel tetrameric complex formed by two dimers of crotoxin B isoforms (CB1 and CB2). The results suggest that these assemblies are stable in solution and show that Ser1 and Glu92 of CB1 and CB2, respectively, play an important role in the oligomerization. The tetrameric and dimeric conformations resulting from the association of the isoforms may increase the neurotoxicity of the toxin CB by the creation of new binding sites, which could improve the affinity of the molecular complexes to the presynaptic membrane.
Organizational Affiliation:
Departamento de Física e Biofísica, Instituto de Biociências, UNESP, Botucatu, SP, Brazil.