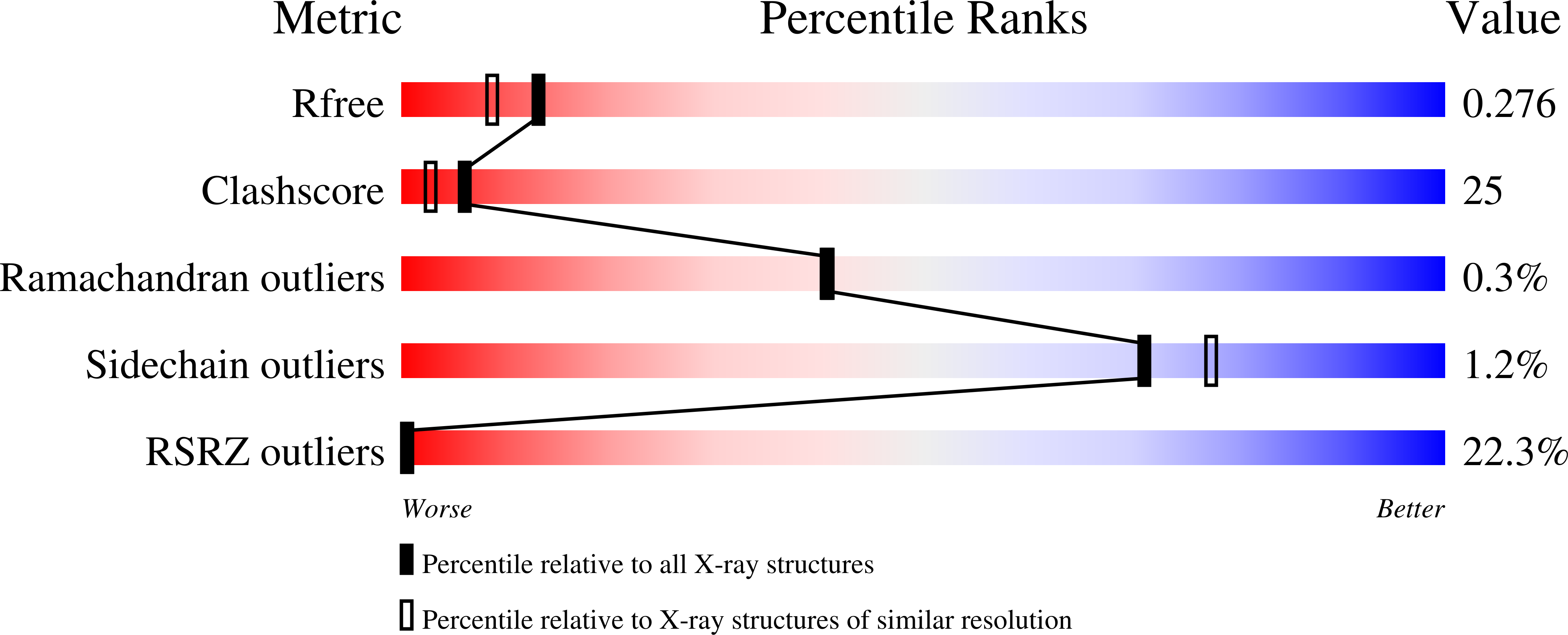
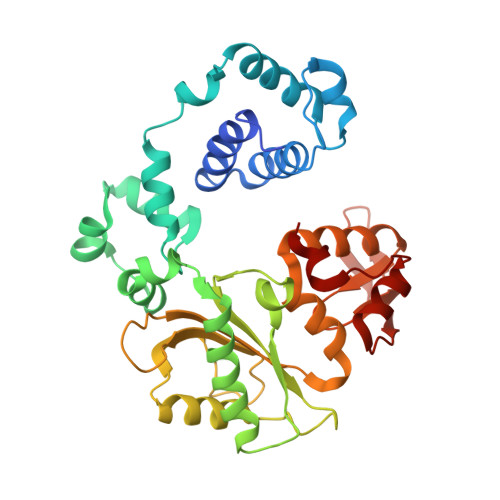
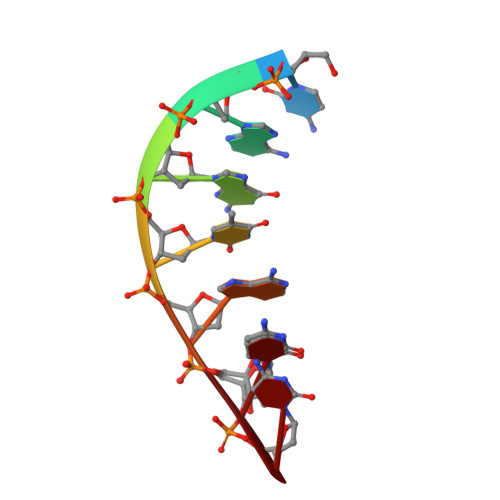
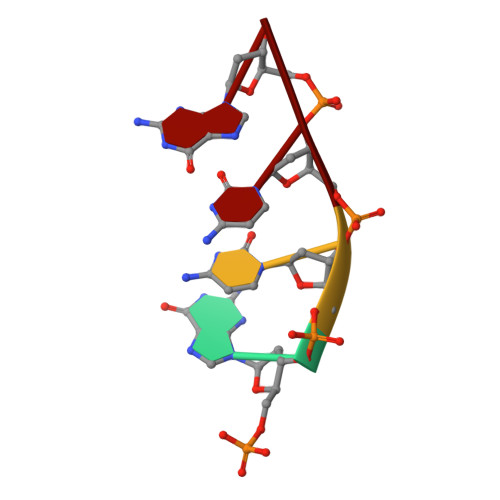
Role of the catalytic metal during polymerization by DNA polymerase lambda.
Garcia-Diaz, M., Bebenek, K., Krahn, J.M., Pedersen, L.C., Kunkel, T.A.(2007) DNA Repair (Amst) 6: 1333-1340
- PubMed: 17475573
- DOI: https://doi.org/10.1016/j.dnarep.2007.03.005
- Primary Citation of Related Structures:
2PFN, 2PFO, 2PFP, 2PFQ - PubMed Abstract:
The incorporation of dNMPs into DNA by polymerases involves a phosphoryl transfer reaction hypothesized to require two divalent metal ions. Here we investigate this hypothesis using as a model human DNA polymerase lambda (Pol lambda), an enzyme suggested to be activated in vivo by manganese. We report the crystal structures of four complexes of human Pol lambda. In a 1.9 A structure of Pol lambda containing a 3'-OH and the non-hydrolyzable analog dUpnpp, a non-catalytic Na+ ion occupies the site for metal A and the ribose of the primer-terminal nucleotide is found in a conformation that positions the acceptor 3'-OH out of line with the alpha-phosphate and the bridging oxygen of the pyrophosphate leaving group. Soaking this crystal in MnCl2 yielded a 2.0 A structure with Mn2+ occupying the site for metal A. In the presence of Mn2+, the conformation of the ribose is C3'-endo and the 3'-oxygen is in line with the leaving oxygen, at a distance from the phosphorus atom of the alpha-phosphate (3.69 A) consistent with and supporting a catalytic mechanism involving two divalent metal ions. Finally, soaking with MnCl2 converted a pre-catalytic Pol lambda/Na+ complex with unreacted dCTP in the active site into a product complex via catalysis in the crystal. These data provide pre- and post-transition state information and outline in a single crystal the pathway for the phosphoryl transfer reaction carried out by DNA polymerases.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, Research Triangle Park, NC 27709, USA.