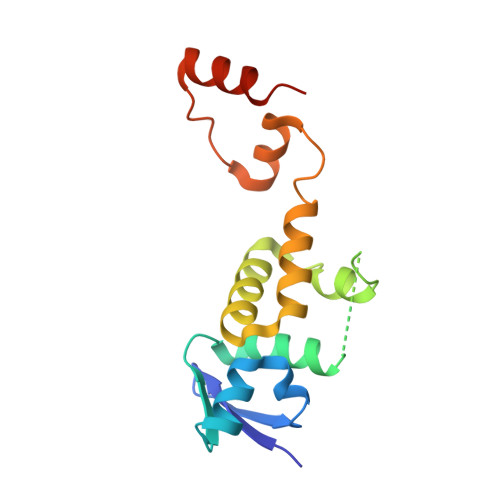
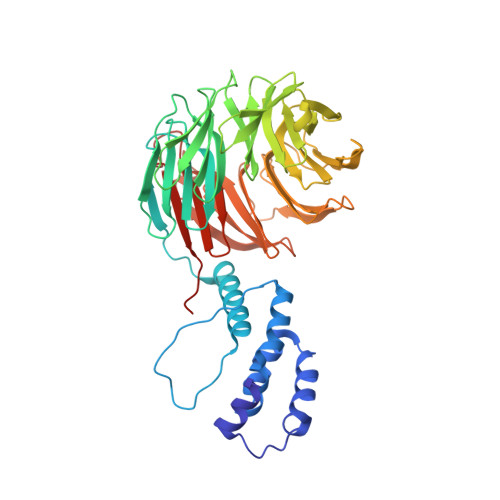
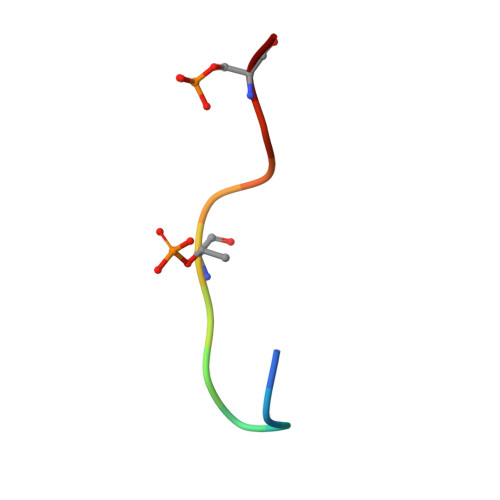
Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Hao, B., Oehlmann, S., Sowa, M.E., Harper, J.W., Pavletich, N.P.(2007) Mol Cell 26: 131-143
- PubMed: 17434132
- DOI: https://doi.org/10.1016/j.molcel.2007.02.022
- Primary Citation of Related Structures:
2OVP, 2OVQ, 2OVR - PubMed Abstract:
The ubiquitin-mediated proteolysis of cyclin E plays a central role in cell-cycle progression, and cyclin E accumulation is a common event in cancer. Cyclin E degradation is triggered by multisite phosphorylation, which induces binding to the SCF(Fbw7) ubiquitin ligase complex. Structures of the Skp1-Fbw7 complex bound to cyclin E peptides identify a doubly phosphorylated pThr380/pSer384 cyclin E motif as an optimal, high-affinity degron and a singly phosphorylated pThr62 motif as a low-affinity one. Biochemical data indicate that the closely related yeast SCF(Cdc4) complex recognizes the multisite phosphorylated Sic1 substrate similarly and identify three doubly phosphorylated Sic1 degrons, each capable of high-affinity interactions with two Cdc4 phosphate binding sites. A model that explains the role of multiple cyclin E/Sic1 degrons is provided by the findings that Fbw7 and Cdc4 dimerize, that Fbw7 dimerization enhances the turnover of a weakly associated cyclin E in vivo, and that Cdc4 dimerization increases the rate and processivity of Sic1 ubiquitination in vitro.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10021, USA.